| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,015,450 – 8,015,557 |
| Length | 107 |
| Max. P | 0.999753 |

| Location | 8,015,450 – 8,015,557 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 54.69 |
| Shannon entropy | 0.94267 |
| G+C content | 0.37010 |
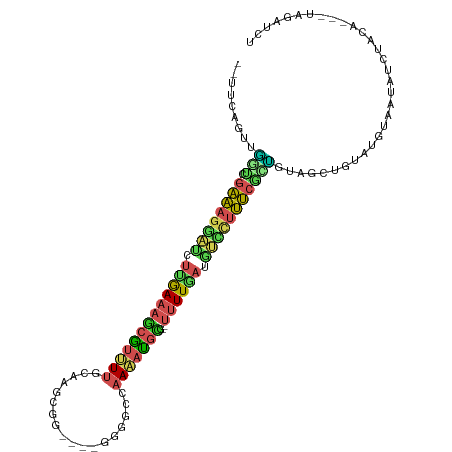
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.76 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
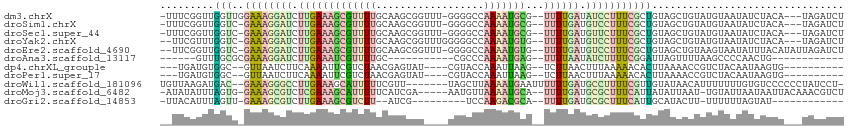
>dm3.chrX 8015450 107 - 22422827 -UUUCGGUUGGUUGGAAAGGAUCUUGAAAGCGUUUUGCAAGCGGUUU-GGGGCCAAAAUGCG--UUUUGAUAUCCUUUCGCUGUAGCUGUAUGUAAUAUCUACA---UAGAUCU -...((((((...(((((((((.(..(((((((((((...((.....-...)))))))))).--)))..).)))))))).)..))))))((((((.....))))---))..... ( -30.50, z-score = -1.83, R) >droSim1.chrX 6409726 106 - 17042790 -UUUCGGUUGGUC-GAAAGGAUCUUGAAAGCGUUUUGCAAGCGGUUU-GGGGCCAAAAUGCG--UUUUGAUGUCCUUUCGCUGUAGCUGUAUGUAAUAUCUACA---UAGAUCU -...((((((..(-((((((((.(..(((((((((((...((.....-...)))))))))).--)))..).)))))))))...))))))((((((.....))))---))..... ( -33.00, z-score = -2.56, R) >droSec1.super_44 101667 106 - 251150 -UUUCGGUUGGUC-GAAAGGAUCUUGAAAGCGUUUUGCAAGCGGUUU-GGGGCCAAAAUGCG--UUUUGAUGUUCUUUCGCUGUAGCUGUAUGUAAUAUCUACA---UAGAUCU -...((((((..(-((((((((.(..(((((((((((...((.....-...)))))))))).--)))..).)))))))))...))))))((((((.....))))---))..... ( -30.10, z-score = -1.83, R) >droYak2.chrX 8475884 106 + 21770863 --UUCGUUUGGUC-GAAAGGAUCUUGAAAGCGUUUUGCAAGCGGUUUGGGGGCCAAAAUGUG--UUUUGAUGUCCUUUCGCUGUAGCUGUAUGUAAUAUCUACA---UAGAUCU --.......((((-((((((((.(..(((((((((((....)((((....))))))))))).--)))..).))))))))..(((((.(((.....))).)))))---..)))). ( -28.80, z-score = -1.57, R) >droEre2.scaffold_4690 10385903 108 - 18748788 --UUCGGUUGGUC-GAAAGGAUCUUGAAAGCGUUUUGCAAGCGGUUU-GGGGCCAAAAUGUG--UUUUGAUGUCCUUUCGCUGUAGCUGUAAGUAAUAUUUACAUAUUAGAUCU --.......((((-((((((((.(..(((((((((((...((.....-...)))))))))).--)))..).))))))))((....))(((((((...))))))).....)))). ( -28.40, z-score = -1.50, R) >droAna3.scaffold_13117 2217140 83 - 5790199 ------GUUUGCGCGAAAGGAUCUUGAAAUCGUUUUGC-----------CGCCCAAAAUGAG--UUUUAAUAUCUUUUCGGAUUAGUUUUAAGCCCCAACUG------------ ------.......(((((((((.((((((((((((((.-----------....)))))))).--)))))).)))))))))......................------------ ( -20.00, z-score = -2.16, R) >dp4.chrXL_group1e 3946329 93 - 12523060 ---UGAUGUGGC--GUUAAUCUUCAAAAUUCGUCUAACGAGUAU----CGUACCAAAUUAAG--UCUUAACUUUAAAAACACUUAAAACCGUCUACAAUAAGUG---------- ---...((((((--(............(((((.....)))))..----.........(((((--(.((.........)).))))))...)).))))).......---------- ( -11.20, z-score = -0.27, R) >droPer1.super_17 881240 93 - 1930428 ---UGAUGUGGC--GUUAAUCUUCAAAAUUCGUCUAACGAGUAU----CGUACCAAAUUAAG--UCUUAACUUUAAAAACACUUAAAACCGUCUACAAUAAGUG---------- ---...((((((--(............(((((.....)))))..----.........(((((--(.((.........)).))))))...)).))))).......---------- ( -11.20, z-score = -0.27, R) >droWil1.scaffold_181096 2190793 104 + 12416693 UGUUAAGAUGAC--GAAAGGGCCUUGAAAGCAUUUUUCGUU-------UAGCUUAAAAUGAAUUUUUUGAUGCCUUUUCGUUGUAUAACAUUUUUUUGUGUCCCCCCUAUCCU- (((((..(..((--((((((((.(..((((.....((((((-------(......))))))).))))..).))))))))))..).))))).......................- ( -26.70, z-score = -4.20, R) >droMoj3.scaffold_6482 1788299 104 + 2735782 -AUAUAUUUAGUG-GAAAGCGUCUCGAAAGCAUUUUUCAUCGA-----AAUGUUAAAAUGCA--UUUUGAUGCGCUUUCAUUAUAUUAAU-UGUAUUAAUAAUUACAAACGUCU -.......(((((-.(((((((.(((((((((((((.(((...-----.)))..))))))).--)))))).))))))))))))......(-((((........)))))...... ( -27.70, z-score = -3.94, R) >droGri2.scaffold_14853 1862183 86 + 10151454 -UUACAUUUAGUU-GAAAGCGUCUUGAAAGCGUCUU--AUCG---------UCCAAGACGCA--UUUUGAUGCGCUUUCAUUGCAUACUU-UUUUUUAGUAU------------ -.........(((-((((((((.(..((((((((((--....---------...))))))).--)))..).)))))))))..))(((((.-......)))))------------ ( -27.80, z-score = -5.01, R) >consensus __UUCAGUUGGUC_GAAAGGAUCUUGAAAGCGUUUUGCAAGCGG____GGGGCCAAAAUGCG__UUUUGAUGUCCUUUCGCUGUAGCUGUAUGUAAUAUCUACA___UAGAUCU ..............((((((((.(((((((((((((..................)))))))...)))))).))))))))................................... (-11.14 = -11.23 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:01 2011