
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,014,993 – 8,015,110 |
| Length | 117 |
| Max. P | 0.549339 |

| Location | 8,014,993 – 8,015,110 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 55.80 |
| Shannon entropy | 0.97713 |
| G+C content | 0.45668 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -6.43 |
| Energy contribution | -6.07 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.89 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
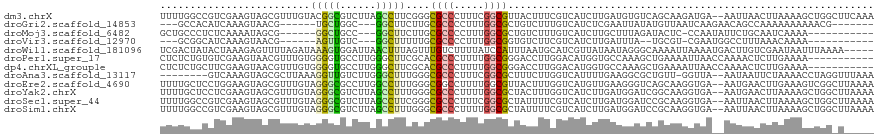
>dm3.chrX 8014993 117 + 22422827 UUUUGGCCGUCGAAGUAGCGUUUGUACGGCGUCUUAGCCUUCGGGCGCCCUUUCGGCGUUACUUUCGUCAUCUUGAUGUGUCAGCAAGAUGA--AAUUAACUUAAAAGCUGGCUUCAAA ....(((((((((((..(((((((...(((......)))..)))))))..)))))))((((.(((((((...((((....))))...)))))--)).)))).........))))..... ( -40.00, z-score = -2.74, R) >droGri2.scaffold_14853 1861802 100 - 10151454 ---GCCACAUCAAAGUAACG------UGCUGGC---GGCUUCUUGCGCCCCUUUGGCGCUGUCUUUGUCAUCUCGAAUUAUAUGUUAAUCAAGAACAGCCAAAAAAAAAACG------- ---(((((((.........)------)).))))---(((((((((((((.....)))))....((((......)))).............))))..))))............------- ( -23.20, z-score = -1.27, R) >droMoj3.scaffold_6482 1787827 98 - 2735782 GCUGCCCUCUCAAAAUAGCG------GGCUGCC---GGCUUCUUGCGCCCCUUUGGCGCUGUCUUUGUCAUCUUGCUUUAGAUACUC-CCAAUAUUCUGCAAUCAAAA----------- ((.((((.((......)).)------))).)).---(((.....(((((.....))))).......)))...((((...(((.....-.......)))))))......----------- ( -23.70, z-score = -0.82, R) >droVir3.scaffold_12970 8331505 93 + 11907090 ---GCGGCAUCAAAGUAACG------AGUUGUC---GGCUUUUUGCGCCCCUUUGGCGGUGUCUUCGUCAUCUUGAUUUA--UGCGU-CGAAUGGCCUUUAAACAAAA----------- ---..((((((...((((.(------(((....---.)))).))))(((.....)))))))))...(((((.(((((...--...))-))))))))............----------- ( -23.90, z-score = -0.26, R) >droWil1.scaffold_181096 2190290 114 - 12416693 UCGACUAUACUAAAGAGUUUUAGAUAAAGUGGAUUAACUUUAGUUUGUCUUUUAUCCAUUUAAUGCAUCGUUAUAAUAGGGCAAAAUUAAAAUGACUUGUCGAAUAAUUUAAAA----- (((((.......(((.(((((((.((((((......)))))).((((((((.........(((((...)))))....)))))))).)))))))..))))))))...........----- ( -16.93, z-score = -0.12, R) >droPer1.super_17 880850 108 + 1930428 CUCUCUGUGUCGAAGUAACGUUUGUGGGGUGCCUUGGGCUUCGCACGCCCUUUUGGCGGGACCUUGGACAUGGUGCCAAAGCUGAAAAUUAACCAAAACUCUUGAAAA----------- .........((.(((....((((.(((...(((..((((.......))))....)))((.(((........))).))...............))))))).)))))...----------- ( -25.80, z-score = 1.26, R) >dp4.chrXL_group1e 3945939 108 + 12523060 CUCUCUGCUUCGAAGUAACGUUUGUGGGGUGCCUUGGGCUUCGCACGCCCUUUUGGCGGGACCUUGGACAUGGUGCCAAAGCUGAAAAUUAACCAAAACUCUUGAAAA----------- ....(..(..((......))...)..)((((((..((((.......))))....)))((.(((........))).))..............)))..............----------- ( -26.60, z-score = 0.98, R) >droAna3.scaffold_13117 2216779 108 + 5790199 --------GUCAAAGUAGCGCUUAAAGGUUGUCUUGGGCUUUGGGCGCCCUUUCGGCGCUUUCUUGGUCAUUUUGAAGGCGCUGUU-GGUUA--AAUAAUUCUAAAACCUAGGUUUAAA --------.........((((((((((.((.....)).))))))))))((...((((((((((...........))))))))))..-))...--..........((((....))))... ( -33.50, z-score = -1.91, R) >droEre2.scaffold_4690 10385447 117 + 18748788 UUUUGCUCCUGGAAGUAGCGUUUGUAGGGCGCCUUGGCCUUUGGGCGGCCUUUUGGCGUUACUUUGGUCAUGUUGAAGGGUCAGCAAGGUGA--AAUGAACUUGAAAGUCGGCUUAAAA ....(((((((.(((.....))).)))).((((((((((....)))(((((((..((((.((....)).))))..)))))))..))))))).--................)))...... ( -39.40, z-score = -1.12, R) >droYak2.chrX 8475443 117 - 21770863 UUUUGCUCCUCGAAGUAGCGUUUGUAGGGCGUCUUAGCCUUUGGGCGCCCUUUUGGCGCUACUUUGGUCAUCUUGAUGGAUCGGCAAGGUGA--AAUGAACUUAAAAGCUGGCUUAAAA ....(((..(((((((((((((...(((((((((........)))))))))...)))))))))))))((((((((.........))))))))--............))).......... ( -45.50, z-score = -3.48, R) >droSec1.super_44 101218 117 + 251150 UUUUGGCCGUCGAAGUAGCGUUUGUAGGGCGUCUUAGCCUUCGGGCGCCCUUUCGGCGCUAUUUUCGUCAUCUUGAUGGAUCCGCAAGGUGA--AAUUAACUUAAAAGCUGGCUUAAAA (((.(((((.((((((((((((...(((((((((........)))))))))...)))))))))))..((((((((.((....))))))))))--.............).))))).))). ( -45.90, z-score = -3.53, R) >droSim1.chrX 6409271 117 + 17042790 UUUUGGCCGUCGAAGUAGCGUUUGUAGGGCGUCUUAGCCUUUGGGCGCCCUUUUGGCGCUAUUUUCGUCAUCUUGAUGGAUCCGCAAGGUGA--AAUUAACUUAAAAGCUGGCUUAAAA (((.(((((.((((((((((((...(((((((((........)))))))))...)))))))))))..((((((((.((....))))))))))--.............).))))).))). ( -47.30, z-score = -4.19, R) >consensus UUUUCCCCGUCGAAGUAGCGUUUGUAGGGUGUCUUAGCCUUCGGGCGCCCUUUUGGCGCUAUCUUCGUCAUCUUGAUGGAUCUGCAAGGUAA__AAUUAACUUAAAAACUGG_U_____ .........................(((((......)))))....((((.....))))............................................................. ( -6.43 = -6.07 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:00 2011