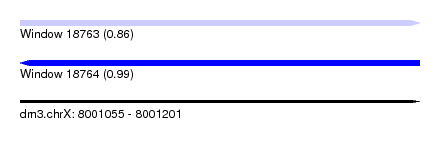
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,001,055 – 8,001,201 |
| Length | 146 |
| Max. P | 0.989434 |

| Location | 8,001,055 – 8,001,201 |
|---|---|
| Length | 146 |
| Sequences | 3 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 86.04 |
| Shannon entropy | 0.18937 |
| G+C content | 0.49322 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -31.71 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
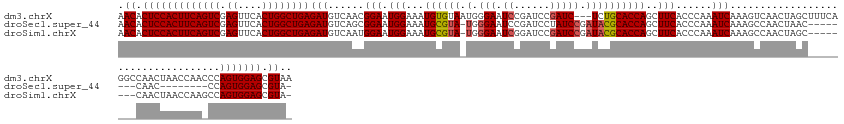
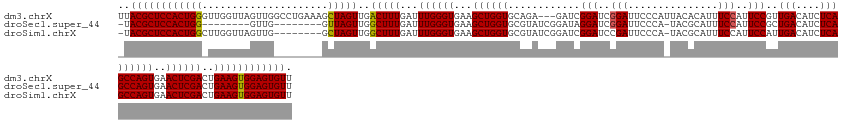
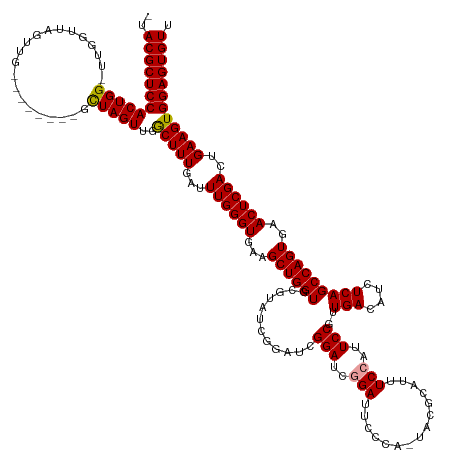
>dm3.chrX 8001055 146 + 22422827 AACACUCCACUUCAGUCGAGUUCACUGGCUGAGAUGUCAACGGAAUGGAAAUGUGUAAUGGGAAUCCGAUCCGAUC---UCUGCACCAGCUUCACCCAAAUCAAAGUCAACUAGCUUUCAGGCCAACUAACCAACCCAGUGGAGCGUAA .((.(((((((((((((.((....)))))))).........((..(((((..(((((((.(((......))).)).---..)))))..(((.....................))))))))..)).............))))))).)).. ( -35.30, z-score = 0.03, R) >droSec1.super_44 84902 131 + 251150 AACACUCCACUUCAGUCGAGUUCACUGGCUGAGAUGUCAGCGGAAUGGAAAUGCGUA-UGGGAAUCCGAUCCUAUCCGAUACGCACCAGCUUCACCCAAAUCAAAGCCAACUAAC--------CAAC--------CCAGUGGAGCGUA- .((.(((((((...((.(.(((...(((((..(((....(.(((.(((...((((((-(.(((...........))).))))))))))..))).)....)))..)))))...)))--------).))--------..))))))).)).- ( -41.80, z-score = -3.05, R) >droSim1.chrX 6396472 139 + 17042790 AACACUCCACUUCAGUCGAGUUCACUGGCUGAGAUGUCAAUGGAAUGGAAAUGCGUA-UGGGAAUCGGAUCCGAUCCGAUACGCACCAGCUUCACCCAAAUCAAAGCCAACUAGC--------CAACUAACCAAGCCAGUGGAGCGUA- .((.(((((((((((((.((....))))))))).......(((..(((...((((((-(.(((.(((....)))))).)))))))...((((...........))))........--------.......)))..))))))))).)).- ( -47.90, z-score = -3.69, R) >consensus AACACUCCACUUCAGUCGAGUUCACUGGCUGAGAUGUCAACGGAAUGGAAAUGCGUA_UGGGAAUCCGAUCCGAUCCGAUACGCACCAGCUUCACCCAAAUCAAAGCCAACUAGC________CAACUAACCAA_CCAGUGGAGCGUA_ .((.(((((((((((((.((....))))))))(((......(((.(((...((((((.(.(((......))).).....)))))))))..)))......)))...................................))))))).)).. (-31.71 = -31.60 + -0.11)
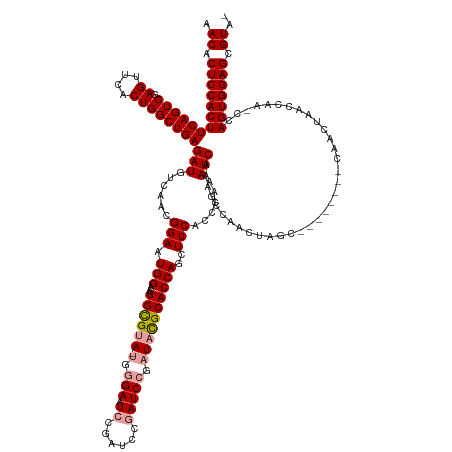
| Location | 8,001,055 – 8,001,201 |
|---|---|
| Length | 146 |
| Sequences | 3 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Shannon entropy | 0.18937 |
| G+C content | 0.49322 |
| Mean single sequence MFE | -55.12 |
| Consensus MFE | -34.91 |
| Energy contribution | -36.47 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8001055 146 - 22422827 UUACGCUCCACUGGGUUGGUUAGUUGGCCUGAAAGCUAGUUGACUUUGAUUUGGGUGAAGCUGGUGCAGA---GAUCGGAUCGGAUUCCCAUUACACAUUUCCAUUCCGUUGACAUCUCAGCCAGUGAACUCGACUGAAGUGGAGUGUU ..((((((((((.(((((((((..((((......))))..))))).......((((...((((((...((---(((((((..(((...............)))..)))).....))))).))))))..))))))))..)))))))))). ( -54.26, z-score = -2.95, R) >droSec1.super_44 84902 131 - 251150 -UACGCUCCACUGG--------GUUG--------GUUAGUUGGCUUUGAUUUGGGUGAAGCUGGUGCGUAUCGGAUAGGAUCGGAUUCCCA-UACGCAUUUCCAUUCCGCUGACAUCUCAGCCAGUGAACUCGACUGAAGUGGAGUGUU -.((((((((((.(--------((((--------(((..((((((..(((.(.((((((...(((((((((.(((...........))).)-))))))))....)).)))).).)))..))))))..))).)))))..)))))))))). ( -55.50, z-score = -4.20, R) >droSim1.chrX 6396472 139 - 17042790 -UACGCUCCACUGGCUUGGUUAGUUG--------GCUAGUUGGCUUUGAUUUGGGUGAAGCUGGUGCGUAUCGGAUCGGAUCCGAUUCCCA-UACGCAUUUCCAUUCCAUUGACAUCUCAGCCAGUGAACUCGACUGAAGUGGAGUGUU -.((((((((((...(..(((((((.--------(((....((((..(((.(.((((((...(((((((((.((((((....))).))).)-))))))))....)).)))).).)))..))))))).)))).)))..))))))))))). ( -55.60, z-score = -3.75, R) >consensus _UACGCUCCACUGG_UUGGUUAGUUG________GCUAGUUGGCUUUGAUUUGGGUGAAGCUGGUGCGUAUCGGAUCGGAUCGGAUUCCCA_UACGCAUUUCCAUUCCGUUGACAUCUCAGCCAGUGAACUCGACUGAAGUGGAGUGUU ..((((((((((((..(((...............(((((((.((((......))))..)))))))(((((.......(((......)))...)))))....)))..)).........(((((.((....)).).)))))))))))))). (-34.91 = -36.47 + 1.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:59 2011