| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,969,483 – 7,969,623 |
| Length | 140 |
| Max. P | 0.872641 |

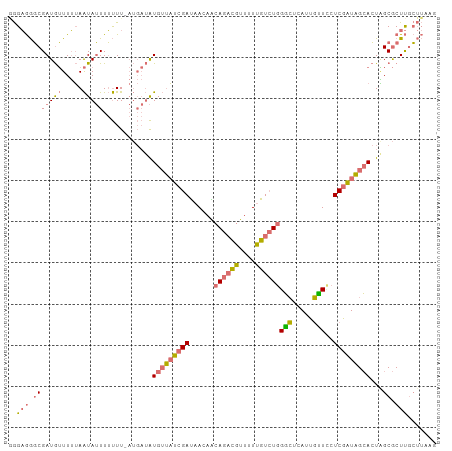
| Location | 7,969,483 – 7,969,586 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.63 |
| Shannon entropy | 0.48138 |
| G+C content | 0.39773 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.85 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
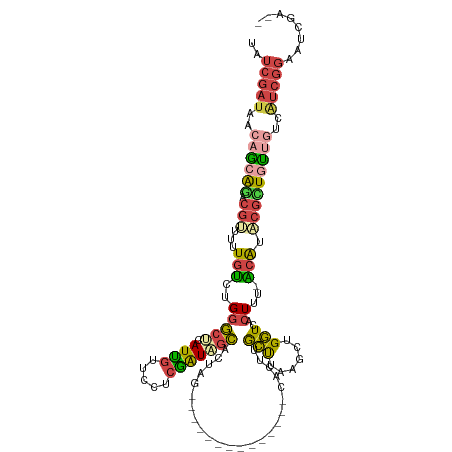
>dm3.chrX 7969483 103 + 22422827 AGGAGGGCGAUGUUUUUAAUAUUUUUUU-AUGAUAUGUUAUCGAUAACAGCAGACGUUUUUGUCUGGGCUCAUUGCUUCUCGAUAGCACUAGCGCUUGCUUAAG (((..((((..((.....(((((.....-..)))))((((((((......((((((....))))))(((.....)))..))))))))))...))))..)))... ( -27.40, z-score = -1.79, R) >droSec1.super_44 53590 104 + 251150 GGGAGGGCGAUGUUUUUAAUAUUUUUUUUAUGAUAUGUUAUCGAUAACAACAGACGUUUUUGUCUGGGCUCAUUGUUCCUCGAUAGCACUAGCGCUUGCUUAAG (.(((.((..........(((((........)))))((((((((.(((((((((((....))))))......)))))..))))))))....)).))).)..... ( -26.10, z-score = -1.90, R) >droSim1.chrX 6366677 104 + 17042790 GGGAGGGCGAUGUUUUUAAUAUUUUUUUUAUGAUAUGUUAUCGAUAAGAACAGACGUUUUUGUCUGGGCUCAUUGUUACUCGAUAGCACUGGCGCUUGCUUAAG (((..((((..((.....(((((........)))))((((((((......((((((....))))))(((.....)))..))))))))))...))))..)))... ( -26.00, z-score = -1.91, R) >droWil1.scaffold_181096 5129834 90 + 12416693 -AGUAAUCGAUAGUUAGAGUAGUUGCUU------GUGAGCUCGACUAUCG-AGCUGUUAUCGU-UGGCGGCAUUUGUCAUCAGUUGGAAUUGCACUCAA----- -.(((((((((((((.((((........------....))))))))))))-)(((((((....-)))))))..................))))......----- ( -24.60, z-score = -0.73, R) >consensus GGGAGGGCGAUGUUUUUAAUAUUUUUUU_AUGAUAUGUUAUCGAUAACAACAGACGUUUUUGUCUGGGCUCAUUGUUCCUCGAUAGCACUAGCGCUUGCUUAAG ..(((.(((((((.....)))))............(((((((((......((((((....))))))(((.....)))..)))))))))...)).)))....... (-14.04 = -14.85 + 0.81)


| Location | 7,969,520 – 7,969,623 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.63250 |
| G+C content | 0.43935 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -10.04 |
| Energy contribution | -9.93 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
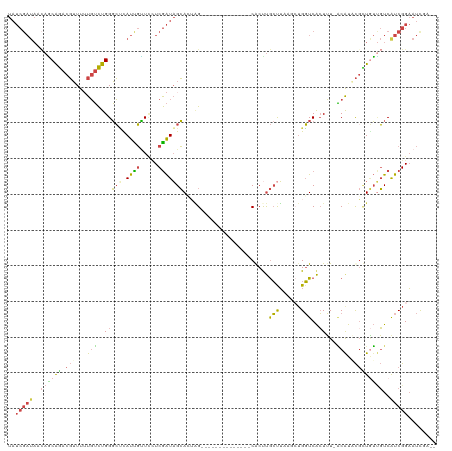
>dm3.chrX 7969520 103 + 22422827 UAUCGAUAACAGCAGACGUUUUUGUCUGGGCUCAUUGCUUCUCGAUAGCACUAG--------------CGCUUGCUUAAGCUGGUCACUUU-ACAUUCGCUGUUGUCAUCGGAAUCUA-- ..(((((.(((((((.((....(((..(((((.((((.....)))))))(((((--------------(..........))))))..))..-)))..))))))))).)))))......-- ( -29.40, z-score = -1.95, R) >droEre2.scaffold_4690 16878651 113 + 18748788 UAUCGAUACCAGCAGACGUUUAUGUCUGGGCCUAUCGUUCUUCGAUAGCAGAAGUUGUAUUUCUAAGCCACCUGCUU----UGGUCACUUU-ACAUACUUUGUUGUCGUCGGAAUCGA-- ..(((((.((..((((((....))))))..............((((((((((.((((((.....(((..(((.....----.)))..))))-))).))))))))))))..)).)))))-- ( -30.50, z-score = -1.69, R) >droYak2.chrX 8428002 118 - 21770863 UAUCGAUACCAGCAGACGUUUUUGUCUGGGCCUAUCGUUCUUCGAUAGCAGAAGGUGUAUUUCACAGUCACUUGCCUCGCCUGGUCACUUUUGCGUUCGAUAUCGUCAUCGGAAUCGA-- ..((((((((((((((((....))))))(((((((((.....))))))..((..(((.....)))..))....)))....)))))..........((((((......)))))))))))-- ( -35.60, z-score = -1.95, R) >droSec1.super_44 53628 105 + 251150 UAUCGAUAACAACAGACGUUUUUGUCUGGGCUCAUUGUUCCUCGAUAGCACUAG--------------CGCUUGCUUAAGCUGGUCACUUU-ACAUACGCUGUUGUCAUCGGAAUUUUAA ..(((((.(((((((.(((...(((..(((((.((((.....)))))))(((((--------------(..........))))))..))..-))).)))))))))).)))))........ ( -32.20, z-score = -3.63, R) >droSim1.chrX 6366715 105 + 17042790 UAUCGAUAAGAACAGACGUUUUUGUCUGGGCUCAUUGUUACUCGAUAGCACUGG--------------CGCUUGCUUAAGCUGGUCACUUU-ACAUACGCUGUUGUCAUCGGAAUUUUUA ..(((((.(.(((((.(((...(((..(((((.((((.....)))))))((..(--------------(..........))..))..))..-))).)))))))).).)))))........ ( -27.30, z-score = -1.53, R) >droWil1.scaffold_181096 5129872 88 + 12416693 ---------UAUCGAGCUGUUAUCGUUGGCGGCAUUUGUCAUCAGUUGGAAUUG--------------CACUCAACG-----GUUCGAUGU-UCAUUUGUUCGAUUCAUCGGU-UCGA-- ---------..((((((....((((((((..(((....(((.....)))...))--------------)..))))))-----)).(((((.-((........))..)))))))-))))-- ( -24.80, z-score = -1.41, R) >consensus UAUCGAUAACAGCAGACGUUUUUGUCUGGGCUCAUUGUUCCUCGAUAGCACUAG______________CACUUGCUUAAGCUGGUCACUUU_ACAUACGCUGUUGUCAUCGGAAUCGA__ ..(((((.....((((((....)))))).(((.((((.....)))))))........................(((......)))......................)))))........ (-10.04 = -9.93 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:55 2011