| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,952,825 – 7,952,901 |
| Length | 76 |
| Max. P | 0.972453 |

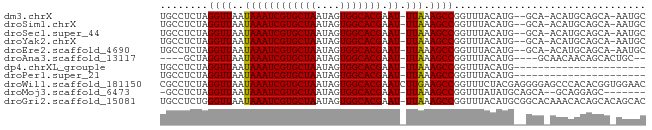
| Location | 7,952,825 – 7,952,901 |
|---|---|
| Length | 76 |
| Sequences | 11 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Shannon entropy | 0.41690 |
| G+C content | 0.43189 |
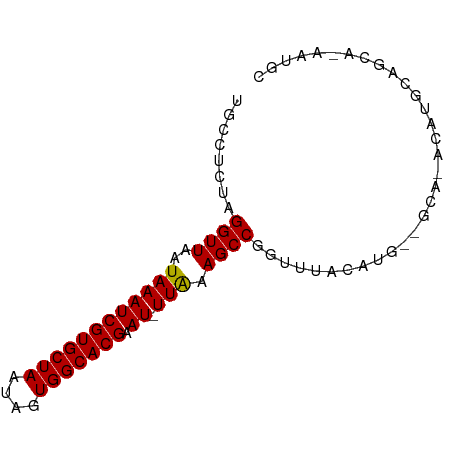
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7952825 76 + 22422827 UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG--GCA-ACAUGCAGCA-AAUGC (((.....((((..((((((((((((....))))))).))-))).))))......((((--...-.))))..)))-..... ( -22.80, z-score = -2.18, R) >droSim1.chrX 6350223 76 + 17042790 UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG--GCA-ACAUGCAGCA-AAUGC (((.....((((..((((((((((((....))))))).))-))).))))......((((--...-.))))..)))-..... ( -22.80, z-score = -2.18, R) >droSec1.super_44 37134 76 + 251150 UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG--GCA-ACAUGCAGCA-AAUGC (((.....((((..((((((((((((....))))))).))-))).))))......((((--...-.))))..)))-..... ( -22.80, z-score = -2.18, R) >droYak2.chrX 8411446 76 - 21770863 UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG--GCA-ACAUGCAGCA-AAUGC (((.....((((..((((((((((((....))))))).))-))).))))......((((--...-.))))..)))-..... ( -22.80, z-score = -2.18, R) >droEre2.scaffold_4690 16861717 76 + 18748788 UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG--GCA-ACAUGCAGCA-AAUGC (((.....((((..((((((((((((....))))))).))-))).))))......((((--...-.))))..)))-..... ( -22.80, z-score = -2.18, R) >droAna3.scaffold_13117 2166465 70 + 5790199 ----GCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG----GCAACAACAGCACUGC-- ----(((..(((..((((((((((((....))))))).))-)))..((((.......))----)))))...))).....-- ( -20.60, z-score = -2.49, R) >dp4.chrXL_group1e 6590316 58 + 12523060 UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG---------------------- .(((....((((..((((((((((((....))))))).))-))).))))))).......---------------------- ( -15.20, z-score = -2.17, R) >droPer1.super_21 1602635 58 + 1645244 UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUG---------------------- .(((....((((..((((((((((((....))))))).))-))).))))))).......---------------------- ( -15.20, z-score = -2.17, R) >droWil1.scaffold_181150 2958510 81 + 4952429 CGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAUCUUGAAGCCGGUUUCUACGAGGGGAGCCCACACGGUGGAAC ((((....((((......((((((((....)))))))).......))))(((((((.....))))))).....)))).... ( -26.72, z-score = -1.88, R) >droMoj3.scaffold_6473 14367696 70 - 16943266 -GCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUAUAUGCAGCA--GCAGGAGC------- -..((((.((((..((((((((((((....))))))).))-))).))))........(((....--))))))).------- ( -20.30, z-score = -1.74, R) >droGri2.scaffold_15081 1042842 80 - 4274704 UGCCUCUGGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU-UUAAAGCCGGUUUACAUGCGGCACAAACACAGCACAGCAC (((..(((.(((..((((((((((((....))))))).))-)))..((((.........))))...))).)))....))). ( -22.60, z-score = -1.71, R) >consensus UGCCUCUAGGUUAAUAAAUCGUGCUAAUAGUGGCACGAAU_UUAAAGCCGGUUUACAUG__GCA_ACAUGCAGCA_AAUGC ........((((......((((((((....)))))))).......))))................................ (-13.90 = -13.90 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:51 2011