
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,875,805 – 7,875,917 |
| Length | 112 |
| Max. P | 0.998257 |

| Location | 7,875,805 – 7,875,917 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 61.77 |
| Shannon entropy | 0.72029 |
| G+C content | 0.49161 |
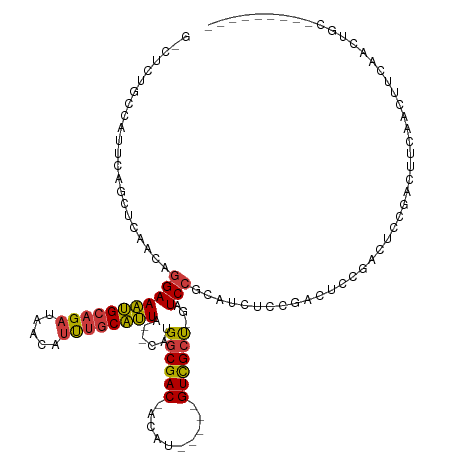
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.04 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
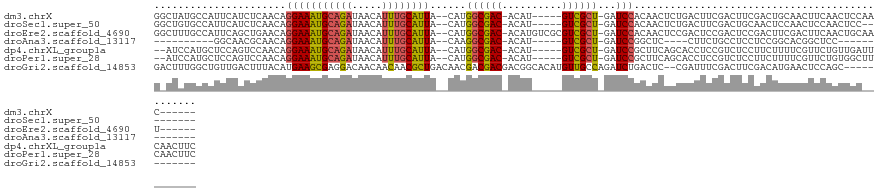
>dm3.chrX 7875805 112 - 22422827 GGCUAUGCCAUUCAUCUCAACAGGAAAUGCAGAUAACAUUUGCAUUA--CAUGGCGAC-ACAU-----GUCGCU-GAUCCACAACUCUGACUUCGACUUCGACUGCAACUUCAACUCCAAC------ (((...))).............((((((((((((...))))))))).--...((((((-....-----))))))-..))).......((...(((....)))...))..............------ ( -24.50, z-score = -2.39, R) >droSec1.super_50 161309 109 - 201529 GGCUGUGCCAUUCAUCUCAACAGGAAAUGCAGAUAACAUUUGCAUUA--CAUGGCGAC-ACAU-----GUCGCU-GAUCCACAACUCUGACUUCGACUGCAACUCCAACUCCAACUCC--------- ((...(((...((.........((((((((((((...))))))))).--...((((((-....-----))))))-..))).((....)).....))..)))...))............--------- ( -23.90, z-score = -1.67, R) >droEre2.scaffold_4690 16788583 117 - 18748788 GGCUUUGCCAUUCAGCUGAACAGGAAAUGCAGAUAACAUUUGCAUUA--CAUGGCGAC-ACAUGUCGCGUCGCU-GAUCCACAACUCCGACUCCGACUCCGACUUCGACUUCAACUGCAAU------ (((...)))...(((.((((..((((((((((((...))))))))).--....(((((-....)))))((((..-(........)..))))))).....((....))..)))).)))....------ ( -29.40, z-score = -1.66, R) >droAna3.scaffold_13117 2094341 91 - 5790199 ----------GGCAACGCAACAGGAAAUGCAGAUAACAUUUGCAUUA--CAAGGCGAC-ACAU-----GUCGCU-GAUCCGGCUC----CUUCUGCCUCCUCCGGCACGGCUCC------------- ----------(((...((...(((((((((((((...))))))))).--...((((((-....-----))))))-.....(((..----.....)))))))...))...)))..------------- ( -29.60, z-score = -2.27, R) >dp4.chrXL_group1a 6732694 116 + 9151740 --AUCCAUGCUCCAGUCCAACAGGAAAUGCAGAUAACAUUUGCAUUA--CAUGGCGAC-ACAU-----GUCGCU-GAUCCGCUUCAGCACCUCCGUCUCCUUCUUUUCGUUCUGUUGAUUCAACUUC --...............(((((((((((((((((...))))))))).--...((((((-....-----)))(((-((......)))))......)))............)))))))).......... ( -29.50, z-score = -2.91, R) >droPer1.super_28 258230 116 + 1111753 --AUCCAUGCUCCAGUCCAACAGGAAAUGCAGAUAACAUUUGCAUUA--CAUGGCGAC-ACAU-----GUCGCU-GAUCCGCUUCAGCACCUCCGUCUCCUUCUUUUCGUUCUGUGGCUUCAACUUC --......((..(((..(....((((((((((((...))))))))).--...((((((-....-----))))))-..)))((....))....................)..)))..))......... ( -29.60, z-score = -2.38, R) >droGri2.scaffold_14853 9314385 113 + 10151454 GACUUUGGCUGUUGACUUUACAUGAAGCGAGGACAACAACAACGCUGACAACGACGACGACGGCACAUGUUGCCAGAUCUGACUC--CGAUUUCGACUUCGACAUGAACUCCAGC------------ .......((((..((.((((..((((((((((.(((((.....((((....((....)).))))...))))))).((((......--.))))))).)))))...)))).))))))------------ ( -21.90, z-score = 0.15, R) >consensus G_CUCUGCCAUUCAGCUCAACAGGAAAUGCAGAUAACAUUUGCAUUA__CAUGGCGAC_ACAU_____GUCGCU_GAUCCGCAUCUCCGACUCCGACUCCGACUUCAACUUCAACUGC_________ ......................(((((((((((.....))))))))......((((((..........))))))...)))............................................... (-14.18 = -14.04 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:43 2011