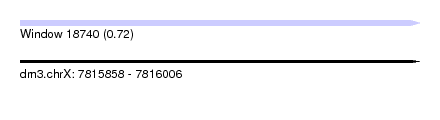
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,815,858 – 7,816,006 |
| Length | 148 |
| Max. P | 0.720708 |

| Location | 7,815,858 – 7,816,006 |
|---|---|
| Length | 148 |
| Sequences | 6 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Shannon entropy | 0.50432 |
| G+C content | 0.51786 |
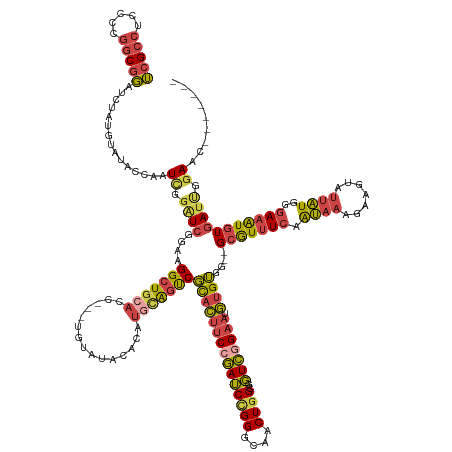
| Mean single sequence MFE | -49.75 |
| Consensus MFE | -32.40 |
| Energy contribution | -33.19 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
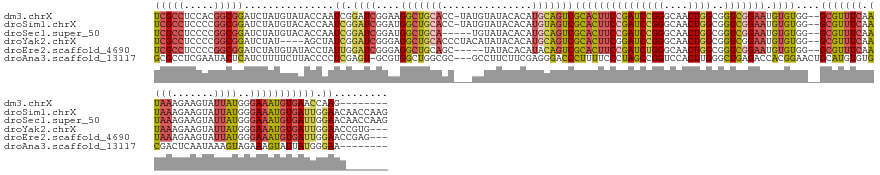
>dm3.chrX 7815858 148 + 22422827 UCGCCUCCACGGCGGAUCUAUGUAUACCAAUCGGAUCGGAAGGCUGCACC-UAUGUAUACACAUGCAGUCGCACUUCCGAUCCGGGCAACUGGCGGUCGGAAUGUGUGG--GCGUUUCAAUAAAGAAGUAUUAUGGGAAAUGUGAACCAAG-------- ..(((.....)))((..(((..(((.((.(((((((((((((((((((..-..(((....)))))))))....))))))))))((....))...))).)).)))..)))--(((((((.((((.......))))..)))))))...))...-------- ( -50.20, z-score = -1.69, R) >droSim1.chrX 6265994 156 + 17042790 UCGCCUCCCCGGCGGAUCUAUGUACACCAAUCGGAUCGGAUGGCUGCACC-UAUGUAUACACAUGUAGUCGCACUUCCGAUCCGGGCAACUGGCGGUCGGAAUGUGUGG--GCGUUUCAAUAAAGAAGUAUUAUGGGAAAUGUGAUUGGAACAACCAAG (((((.....)))))...........(((((((.(((((....)))..((-((((.((((..((((..(((((((((((((((((....))))..))))))).))))))--))))(((......)))))))))))))..)).))))))).......... ( -50.40, z-score = -0.69, R) >droSec1.super_50 102850 152 + 201529 UCGCCUCCCCGGCGGAUCUAUGUACACCAAUCGGAUCGGAUGGCUGCA-----UGUAUACACAUGCAGUCGCACUUCCGAUCCGGGCAACUGGCGGUCGGAAUGUGUGG--GCGUUUCAAUAAAGAAGUAUUAUGGGAAAUGUGAUUGGAACAACCAAG (((((.....)))))...........((((((.........(((((((-----(((....))))))))))(((((((((((((((....))))..))))))).))))..--(((((((.((((.......))))..))))))))))))).......... ( -58.10, z-score = -2.79, R) >droYak2.chrX 8273663 150 - 21770863 UCGCCUCCCCGGCGGAUCUAU----AGCUAUCGGAUCGGGAGGCUGCACCCUACAUAUACACAUGCAGUCGCACUUCGGAUCCGGGCAACUGGCGGUCGGAAUGUGUGG--GCGUUUCAAUAAAGAAGUAUUAUGGGAAAUGUGAUUGGAACCGUG--- ..(((((((...(.(((....----....))).)...)))))))....(((.....((((..((((...((((((((.(((((((....))))..))).))).))))).--))))(((......)))))))...)))...................--- ( -47.40, z-score = 0.38, R) >droEre2.scaffold_4690 16727928 149 + 18748788 UCGCCUCCCCGGCGGAUCUAUGUAUACCUAUUGGAUCGGGAGGCUGCAGC-----UAUACACAUACAGUCGCACUUCCGAUCUGGGCAACUGGCGGUCGGAAUGUGUGG--GCGUUUCAAUAAAGAAGUAUUAUGGGAAAUGUGAUUGGAACCGAG--- (((((((((.....((((((...........)))))))))))))......-----..........(((((((((((((((((..(....)..)..))))))).))))..--(((((((.((((.......))))..)))))))))))).....)).--- ( -49.50, z-score = -1.05, R) >droAna3.scaffold_13117 800022 147 + 5790199 GCGCCUCGAAUACUCAUCUUUUCUUACCCCUCGAGU-GCGUGGCUGGCGC---GCCUUCUUCGAGGGACCCUUUUCCCUAGCCGGUCCACUUGGGCUGAGACCACGGAACUGCAUGUGUGCGACUCAAUAAAGUAGAAAGUAUUAUGGGAA-------- ...(((..((((((..(((...(((....(((((((-(...(((((((.(---(.......))(((((......))))).))))))))))))))).((((.(((((.(......).)))).).))))...))).))).))))))..)))..-------- ( -42.90, z-score = 0.35, R) >consensus UCGCCUCCCCGGCGGAUCUAUGUAUACCAAUCGGAUCGGAAGGCUGCACC___UGUAUACACAUGCAGUCGCACUUCCGAUCCGGGCAACUGGCGGUCGGAAUGUGUGG__GCGUUUCAAUAAAGAAGUAUUAUGGGAAAUGUGAUUGGAAC_______ (((((.....)))))...............((.((((....(((((((...............)))))))(((((((((((((((....))))..))))))).))))....(((((((.((((.......))))..))))))))))).))......... (-32.40 = -33.19 + 0.79)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:38 2011