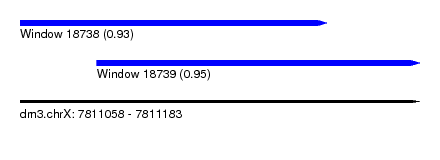
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,811,058 – 7,811,183 |
| Length | 125 |
| Max. P | 0.953657 |

| Location | 7,811,058 – 7,811,154 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.78 |
| Shannon entropy | 0.60570 |
| G+C content | 0.64243 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -20.87 |
| Energy contribution | -20.63 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
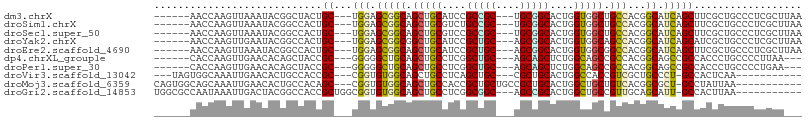
>dm3.chrX 7811058 96 + 22422827 ------AACCAAGUUAAAUACGGCUACUGC---UGGAGCGGCAGCUGCAUCCGCCGC---UGCGGCACUGGUGGCUGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAA ------....((((......((((....))---))(((.((((((.((....((((.---.(((((.(....))))))..))))....))...))))))))))))).. ( -39.60, z-score = -1.01, R) >droSim1.chrX 6261119 96 + 17042790 ------AACCAAGUUAAAUACGGCCACUGC---UGGAGCGGCAGCUGCGUCUGCCGC---UGCGGCACUGGUGGCUGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAA ------....((((......((((((((((---((.((((((((......)))))))---).))))...))))))))....((((.(......).))))...)))).. ( -41.80, z-score = -1.25, R) >droSec1.super_50 97982 96 + 201529 ------AACCAAGUUAAAUACGGCCACUGC---UGGAGCGGCAGCUGCGUCCGCCGC---UGCGGCACUGGUGGCUGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAA ------....((((......((((....))---))(((.((((((.((....((((.---.(((((.(....))))))..))))....))...))))))))))))).. ( -39.50, z-score = -0.45, R) >droYak2.chrX 8268865 96 - 21770863 ------AACCAAGUUGAAUACGGCCACUGC---UGGAGCGGCGGCUGCAUCCGCUGC---AGCGGCACUGGUGGCAGCCACGGCAUCAGCAUCGCUGCCCUCGCUUAA ------....((((.((.....((((.(((---((..(((((((......)))))))---..))))).))))((((((....((....))...)))))).)))))).. ( -40.50, z-score = -0.52, R) >droEre2.scaffold_4690 16722939 96 + 18748788 ------AACCAAGUUAAAUACGGCCACUGC---UGGAGCGGCAGCUGCAUCCGCUGC---AGCGGCACUGGUGGCGGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAA ------....((((......((((....))---))(((.((((((.....((((...---.))))..((((((.((....)).))))))....))))))))))))).. ( -39.20, z-score = -0.48, R) >dp4.chrXL_group1e 12131374 93 - 12523060 ------CACCAAGUUGAACACAGCUACCGC---GGGGGCUGCAGCUGCCUCGGCUGC---AGCAGCUCUGGCAGCCGCCACGGCAGCCGCCACCCUGCCCCUUAA--- ------.....(((((....)))))...((---((((((((((((((...)))))))---))).....((((.((.((....)).)).)))))))))).......--- ( -45.40, z-score = -2.17, R) >droPer1.super_30 828521 93 + 958394 ------CACCAAGUUGAACACAGCUACCGC---GGGGGCUGCAGCUGCCUCGGCUGC---AGCAGCUCUGGCAGCCGCCACGGCAGCCGCCACCCUGCCCCUGAA--- ------.....(((((....)))))...((---((((((((((((((...)))))))---))).....((((.((.((....)).)).)))))))))).......--- ( -45.40, z-score = -1.73, R) >droVir3.scaffold_13042 3026897 87 - 5191987 ---UAGUGGCAAAUUGAACACUGCCACCGC---CGGUGUGGCAGCUGCCUCAGCUGC---CGCUGCACUGGCCACCGUCGCUGCCCU-GCCACUCAA----------- ---..((((((..........)))))).((---((((((((((((((...)))))))---)...))))))))....((.((......-)).))....----------- ( -37.90, z-score = -2.24, R) >droMoj3.scaffold_6359 2458898 93 - 4525533 CAGUGGCAGCAAAUUGAACACUGCCACAGC---CGGUGUGGCAGCUGCCACCGCUGCUGCCGCUGCACUGGCUGCUGUCACGGCGCU-GCCUAUUAA----------- ....((((((.................(((---((((((((((((.((....)).))))))...)))))))))((((...)))))))-)))......----------- ( -44.30, z-score = -1.74, R) >droGri2.scaffold_14853 6694777 93 - 10151454 UGGCGCCAAUAAAUUGACUACGGCCACCGCUGGCGGUGUGGCAGCUGCCUCGGCGGC---AGCCGCACUGGCUGCCGUUGCAGCAUU-GCCACUUAA----------- ((((((((.........(....).(((((....))))))))).(((((...((((((---((((.....)))))))))))))))...-)))).....----------- ( -47.60, z-score = -2.44, R) >consensus ______AACCAAGUUGAAUACGGCCACUGC___UGGAGCGGCAGCUGCCUCCGCUGC___AGCGGCACUGGCGGCCGCCACGGCAUCAGCCUCGCUGCCCUCGCU___ ......................(((.((......)).(.(((.(((((....(((((....)))))....))))).))).))))........................ (-20.87 = -20.63 + -0.24)

| Location | 7,811,082 – 7,811,183 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.00 |
| Shannon entropy | 0.69714 |
| G+C content | 0.65044 |
| Mean single sequence MFE | -45.97 |
| Consensus MFE | -20.22 |
| Energy contribution | -19.67 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
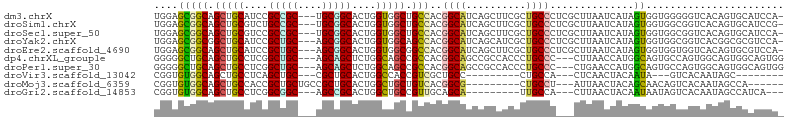
>dm3.chrX 7811082 101 + 22422827 UGGAGCGGCAGCUGCAUCCGCCGC---UGCGGCACUGGUGGCUGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAAUCAUAGUGGUGGGGGUCACAGUGCAUCCA- .(((...(((((.((....)).))---))).((((((.(((((.((((((((.(......).))))..((((.......)))))))).))))))))))).))).- ( -48.10, z-score = -1.40, R) >droSim1.chrX 6261143 101 + 17042790 UGGAGCGGCAGCUGCGUCUGCCGC---UGCGGCACUGGUGGCUGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAAUCAUAGUGGUGGCGGUCACAGUGCAUCCG- .(.((((((((......)))))))---).).((((((.((((((((((((((.(......).))))..((((.......)))))))))))))))))))).....- ( -53.50, z-score = -2.94, R) >droSec1.super_50 98006 101 + 201529 UGGAGCGGCAGCUGCGUCCGCCGC---UGCGGCACUGGUGGCUGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAAUCAUAGUGGUGGCGGUCACAGUGCAUCCA- .(((...(((((.((....)).))---))).((((((.((((((((((((((.(......).))))..((((.......)))))))))))))))))))).))).- ( -52.40, z-score = -2.47, R) >droYak2.chrX 8268889 101 - 21770863 UGGAGCGGCGGCUGCAUCCGCUGC---AGCGGCACUGGUGGCAGCCACGGCAUCAGCAUCGCUGCCCUCGCUUAAUCAUAGUGGUGGCGGUCACGGCGCGUCCA- .((((((((.((((((.....)))---))).))....(((((.(((((((((.(......).))))..((((.......))))))))).)))))..))).))).- ( -47.60, z-score = -0.61, R) >droEre2.scaffold_4690 16722963 101 + 18748788 UGGAGCGGCAGCUGCAUCCGCUGC---AGCGGCACUGGUGGCGGCCACGGCAUCAGCUUCGCUGCCCUCGCUUAAUCAUAGUGGUGGUGGUCACAGUGCGUCCA- .(((((.(((((.......)))))---.)).((((((.((((.(((((((((.(......).))))..((((.......))))))))).)))))))))).))).- ( -48.20, z-score = -1.37, R) >dp4.chrXL_group1e 12131398 99 - 12523060 GGGGGCUGCAGCUGCCUCGGCUGC---AGCAGCUCUGGCAGCCGCCACGGCAGCCGCCACCCUGCCC---CUUAACCAUGGCAGUGCCAGUGGCAGUGGCAGUGG ((.((((((.((((((..(((((.---..)))))..))))))((...)))))))).)).(((((((.---((...(((((((...)))).))).)).))))).)) ( -52.70, z-score = -0.19, R) >droPer1.super_30 828545 99 + 958394 GGGGGCUGCAGCUGCCUCGGCUGC---AGCAGCUCUGGCAGCCGCCACGGCAGCCGCCACCCUGCCC---CUGAACCAUGGCAGUGCCAGUGGCAGUGGCAGUGG ((.((((((.((((((..(((((.---..)))))..))))))((...)))))))).)).(((((((.---(((..(((((((...)))).)))))).))))).)) ( -55.40, z-score = -0.60, R) >droVir3.scaffold_13042 3026924 79 - 5191987 CGGUGUGGCAGCUGCCUCAGCUGC---CGCUGCACUGGCCACCGUCGCUGCC---------CUGCCA---CUCAACUACAAUA---GUCACAAUAGC-------- .(((((((((((((...)))))))---))).(((.((((....)))).))).---------..))).---.............---...........-------- ( -26.80, z-score = -0.56, R) >droMoj3.scaffold_6359 2458928 87 - 4525533 CGGUGUGGCAGCUGCCACCGCUGCUGCCGCUGCACUGGCUGCUGUCACGGCG---------CUGCCU---AUUAACUACAGCAACAGUCACAAUAGCCA------ .(((((((((((.((....)).))))))))((.((((..((((((...(((.---------..))).---.......)))))).)))))).....))).------ ( -34.30, z-score = -0.32, R) >droGri2.scaffold_14853 6694810 87 - 10151454 CGGUGUGGCAGCUGCCUCGGCGGC---AGCCGCACUGGCUGCCGUUGCAGCA---------UUGCCA---CUUAACUACAAUAAUAGUCACAAUAGCCAUCA--- .((((((((((.(((..(((((((---((((.....)))))))))))..)))---------))))))---)...((((......)))).......)))....--- ( -40.70, z-score = -3.92, R) >consensus UGGAGCGGCAGCUGCCUCCGCUGC___AGCGGCACUGGCGGCCGCCACGGCAUCAGCUUCGCUGCCC___CUUAACCAUAGUAGUGGCGGUCACAGUGCAUCCA_ ....(((((.(((((....(((((....)))))....))))).)))..((((..........))))..............))....................... (-20.22 = -19.67 + -0.55)
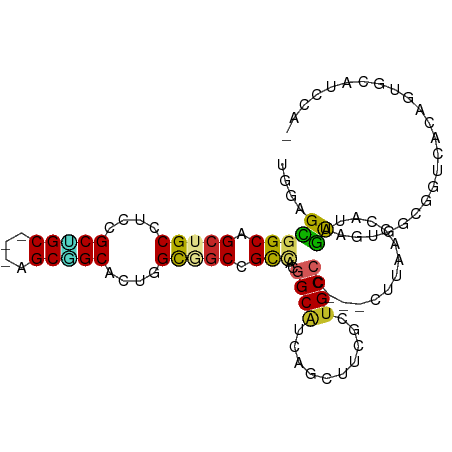
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:37 2011