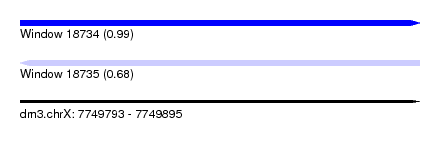
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,749,793 – 7,749,895 |
| Length | 102 |
| Max. P | 0.991750 |

| Location | 7,749,793 – 7,749,895 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 57.20 |
| Shannon entropy | 0.88646 |
| G+C content | 0.60036 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -6.17 |
| Energy contribution | -6.49 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991750 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
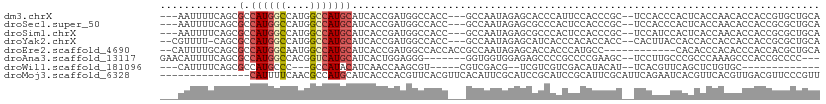
>dm3.chrX 7749793 102 + 22422827 ---AAUUUUCAGCGCCAUGGCCAUGGCCAUGCAUCACCGAUGGCCACC---GCCAAUAGAGCACCCAUUCCACCCGC--UCCACCCACUCACCAACACCACCGUGCUGCA ---......((((((..((((..(((((((.(......))))))))..---))))...((((.............))--)).....................)))))).. ( -29.52, z-score = -3.58, R) >droSec1.super_50 37371 102 + 201529 ---AAUUUUCAGCGCCAUGGCCAUGGCCAUGCAUCACCGAUGGCCACC---GCCAAUAGAGCGCCCACUCCACCCGC--UCCACCCACUCACCAACACCACCGCGCUGCA ---......((((((..((((..(((((((.(......))))))))..---))))...(((((...........)))--)).....................)))))).. ( -33.20, z-score = -4.54, R) >droSim1.chrX 6201113 102 + 17042790 ---AAUUUUCAGCGCCAUGGCCAUGGCCAUGCAUCACCGAUGGCCACC---GCCAAUAGAGCGCCCACUCCACCCGC--UCCAUCCACUCACCAACACCACCGCGCUGCA ---......((((((..((((..(((((((.(......))))))))..---))))...(((((...........)))--)).....................)))))).. ( -33.20, z-score = -4.33, R) >droYak2.chrX 8207393 102 - 21770863 --CGUUUU-CAGCGCCAUGGCCAUGGCCAUGCAUCACCGAUGGCCACC---GCCAAUAGAGCAUCACCCACACCACC--CACUUACCACCACCACCACCACCGCGCUGCA --......-((((((..((((..(((((((.(......))))))))..---))))...(.....)............--.......................)))))).. ( -26.20, z-score = -2.73, R) >droEre2.scaffold_4690 16662646 96 + 18748788 --CAUUUUGCAGCGCCAUGGCAAUGGCCAUGCAUCACCGAUGGCCACCACCGCCAAUAGAGCACCACCCAUGCC------------CACACCCACACCCACCACGCUGCA --.....(((((((...((((..(((((((.(......)))))))).....))))...(.(((.......))).------------)................))))))) ( -27.80, z-score = -2.70, R) >droAna3.scaffold_13117 4190330 98 - 5790199 GAACAUUUUCAGCGCCAUGGCCACGGUCAUGCAUCACUGGAGGG-------GGUGGUGGAGAGCCCCGCCCCGAAGC--UCCUUGCCCGCCCAAAGCCCACCGCCCC--- ......((((((.(.((((((....))))))...).))))))((-------((((((((.((((..((...))..))--)).......((.....))))))))))))--- ( -38.40, z-score = -0.95, R) >droWil1.scaffold_181096 2697658 82 - 12416693 ---CAUUUUCAGCGCCAUGCCC---GCCAUACAUCAACCAAGCGU-----CGUCGACG--UCGUCGUCGACAUACAU--UCACGUUCAGCUCUGUGC------------- ---........((((...((.(---((..............))).-----.(((((((--....)))))))......--.........))...))))------------- ( -18.74, z-score = -1.28, R) >droMoj3.scaffold_6328 1038015 95 + 4453435 ---------------CAUUUUCAACGCCAUGCAUCACCCACGUUCACGUUCACAUUCGCAUCCGCAUCCGCAUUCGCAUUCAGAAUCACGUUCACGUUGACGUUCCCGUU ---------------.......((((....((.(((...((((..((((....((((((....))....((....)).....)))).))))..))))))).))...)))) ( -14.70, z-score = -1.07, R) >consensus ___AAUUUUCAGCGCCAUGGCCAUGGCCAUGCAUCACCGAUGGCCACC___GCCAAUAGAGCGCCCACCCCACCCGC__UCCAUCCCACCACCAACACCACCGCGCUGCA ...............((((((....))))))............................................................................... ( -6.17 = -6.49 + 0.31)
| Location | 7,749,793 – 7,749,895 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 57.20 |
| Shannon entropy | 0.88646 |
| G+C content | 0.60036 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -5.13 |
| Energy contribution | -5.92 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.13 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680550 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 7749793 102 - 22422827 UGCAGCACGGUGGUGUUGGUGAGUGGGUGGA--GCGGGUGGAAUGGGUGCUCUAUUGGC---GGUGGCCAUCGGUGAUGCAUGGCCAUGGCCAUGGCGCUGAAAAUU--- ..((((.(.(((((........((.((((((--(((...........))))))))).))---.((((((((((....)).)))))))).))))).).))))......--- ( -41.00, z-score = -2.02, R) >droSec1.super_50 37371 102 - 201529 UGCAGCGCGGUGGUGUUGGUGAGUGGGUGGA--GCGGGUGGAGUGGGCGCUCUAUUGGC---GGUGGCCAUCGGUGAUGCAUGGCCAUGGCCAUGGCGCUGAAAAUU--- ..((((((.(((((........((.((((((--(((...........))))))))).))---.((((((((((....)).)))))))).))))).))))))......--- ( -48.50, z-score = -3.80, R) >droSim1.chrX 6201113 102 - 17042790 UGCAGCGCGGUGGUGUUGGUGAGUGGAUGGA--GCGGGUGGAGUGGGCGCUCUAUUGGC---GGUGGCCAUCGGUGAUGCAUGGCCAUGGCCAUGGCGCUGAAAAUU--- ..((((((.(((((........((.((((((--(((...........))))))))).))---.((((((((((....)).)))))))).))))).))))))......--- ( -49.00, z-score = -4.06, R) >droYak2.chrX 8207393 102 + 21770863 UGCAGCGCGGUGGUGGUGGUGGUGGUAAGUG--GGUGGUGUGGGUGAUGCUCUAUUGGC---GGUGGCCAUCGGUGAUGCAUGGCCAUGGCCAUGGCGCUG-AAAACG-- ..((((((.(((((.(((((((((((..((.--(((((.(((.....))).))))).))---....)))))).(((...))).))))).))))).))))))-......-- ( -45.80, z-score = -3.23, R) >droEre2.scaffold_4690 16662646 96 - 18748788 UGCAGCGUGGUGGGUGUGGGUGUG------------GGCAUGGGUGGUGCUCUAUUGGCGGUGGUGGCCAUCGGUGAUGCAUGGCCAUUGCCAUGGCGCUGCAAAAUG-- ((((((((.((....((.((((.(------------(((((.....)))))))))).))((..((((((((((....)).))))))))..)))).)))))))).....-- ( -48.20, z-score = -3.83, R) >droAna3.scaffold_13117 4190330 98 + 5790199 ---GGGGCGGUGGGCUUUGGGCGGGCAAGGA--GCUUCGGGGCGGGGCUCUCCACCACC-------CCCUCCAGUGAUGCAUGACCGUGGCCAUGGCGCUGAAAAUGUUC ---((((.(((((((((.....))))..(((--((((((...)))))))))))))).))-------))...(((((...((((.(....).)))).)))))......... ( -41.80, z-score = 0.48, R) >droWil1.scaffold_181096 2697658 82 + 12416693 -------------GCACAGAGCUGAACGUGA--AUGUAUGUCGACGACGA--CGUCGACG-----ACGCUUGGUUGAUGUAUGGC---GGGCAUGGCGCUGAAAAUG--- -------------...(((.(((........--..((.((((((((....--))))))))-----))(((((.(........).)---))))..))).)))......--- ( -25.10, z-score = -0.73, R) >droMoj3.scaffold_6328 1038015 95 - 4453435 AACGGGAACGUCAACGUGAACGUGAUUCUGAAUGCGAAUGCGGAUGCGGAUGCGAAUGUGAACGUGAACGUGGGUGAUGCAUGGCGUUGAAAAUG--------------- ..((....))(((((((.(.(((.((((....((((..(((....)))..)))).......(((....))))))).)))..).))))))).....--------------- ( -24.00, z-score = -0.34, R) >consensus UGCAGCGCGGUGGUGUUGGUGAGGGGAUGGA__GCGGGUGGGGGGGGCGCUCUAUUGGC___GGUGGCCAUCGGUGAUGCAUGGCCAUGGCCAUGGCGCUGAAAAUU___ .......................................................................(((((...((((........)))).)))))......... ( -5.13 = -5.92 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:33 2011