
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,749,412 – 7,749,493 |
| Length | 81 |
| Max. P | 0.987201 |

| Location | 7,749,412 – 7,749,493 |
|---|---|
| Length | 81 |
| Sequences | 9 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.53413 |
| G+C content | 0.47337 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -12.38 |
| Energy contribution | -12.59 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
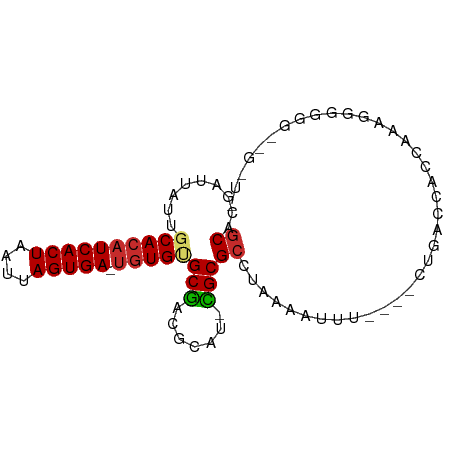
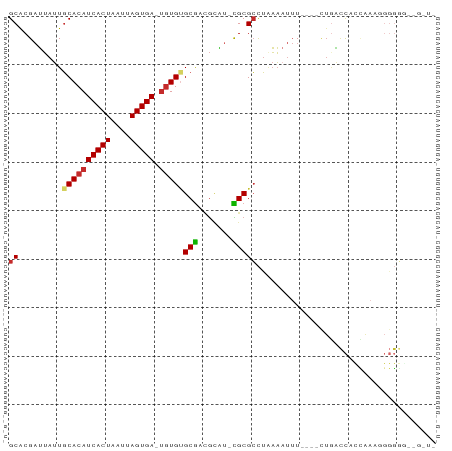
>dm3.chrX 7749412 81 + 22422827 GCACGAUUAUUGCACAUCACUAAUUAGUGA-UGUGUGCGACGCAU-CGCGCCUAAAAUUU----CUGACCACCAAAAGGGGGA-GGUG ((.........((((((((((....)))))-)))))((((....)-))))).........----((..((.((....))))..-)).. ( -25.80, z-score = -2.46, R) >droAna3.scaffold_13117 4189649 82 - 5790199 GCACGAUUAUUGCACAUCACUAAUUAGUGA-UGUGUGCGACGCAU-CGCGCCUGAAAUUU----CUAACCAUCAAAUUGGGGCGGAUG ...........((((((((((....)))))-)))))......(((-(.((((.((....)----)...(((......))))))))))) ( -26.30, z-score = -2.54, R) >droEre2.scaffold_4690 16662256 80 + 18748788 GCACGAUUAUUGCACAUCACUAAUUAGUGA-UGUGUGCGACGCAU-CGCGCCUAAAAUUU----CUGACCAUUAAAGGGGGA--GGUG ((.........((((((((((....)))))-)))))((((....)-))))).....((((----((..((......)).)))--))). ( -25.40, z-score = -2.58, R) >droYak2.chrX 8206986 82 - 21770863 GCACGAUUAUUGCACAUCACUAAUUAGUGA-UGUGUGCGACGCAU-CGCGCCUAAAAUUU----CUGACCAUCAAAAGGGGGAAGGUG ...........((((((((((....)))))-)))))((((....)-)))((((....(..----((..........))..)..)))). ( -24.10, z-score = -1.98, R) >droSim1.chrX 6200750 76 + 17042790 GCACGAUUAUUGCACAUCACUAAUUAGUGA-UGUGUGCGACGCAU-CGCGCCUAAAAUUU----CUGACCACCAAAGGGGGA------ ((.........((((((((((....)))))-)))))((((....)-))))).........----....((.((....)))).------ ( -23.60, z-score = -2.44, R) >droSec1.super_50 37044 75 + 201529 GCACGAUUAUUGCACAUCACUAAUUAGUGA-UGUGUGCGACGCAU-CGCGCCUAAAAUUU----CUGACCACCAAAGGGGG------- ((.........((((((((((....)))))-)))))((((....)-))))).........----....((.((....))))------- ( -22.50, z-score = -2.30, R) >droWil1.scaffold_181096 2697353 64 - 12416693 GCACGAUUAUUGCACAUCACUAAUUAGUGAAUGUGCGCGUGGUAU-UGCGCUU-AAAUUU----CAGAUU------------------ (((..((((((((((((((((....)))).))))))).)))))..-)))....-......----......------------------ ( -18.00, z-score = -1.81, R) >droVir3.scaffold_13042 2551560 87 + 5191987 GCACGAUUAUUGCACAUCACUAAUUAGUGA-UGUGAGCGCCGCGCGCGCGCCUAAAUUUUACGUUUCGGCACGAAUAGAUUGCGGGUC (((...(((((.(((((((((....)))))-)))).((((.....))))(((.((((.....)))).)))...)))))..)))..... ( -27.20, z-score = -0.72, R) >droMoj3.scaffold_6328 3416053 76 - 4453435 GCGCGCGGAAUUCACAUCACUAAUUAGUGA---UGUGCAAUAAUCGUGCGGCAACUGCAUG---AUGCCCAUCGAGGGGGCG------ .(((((((.((((((((((((....)))))---)))).)))..)))))))((....))...---..((((.......)))).------ ( -31.50, z-score = -3.13, R) >consensus GCACGAUUAUUGCACAUCACUAAUUAGUGA_UGUGUGCGACGCAU_CGCGCCUAAAAUUU____CUGACCACCAAAGGGGGG__G_U_ ((.........((((((((((....))))).)))))(((.......)))))..................................... (-12.38 = -12.59 + 0.21)

| Location | 7,749,412 – 7,749,493 |
|---|---|
| Length | 81 |
| Sequences | 9 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.53413 |
| G+C content | 0.47337 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
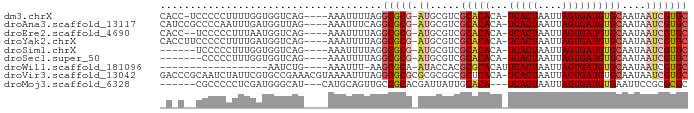
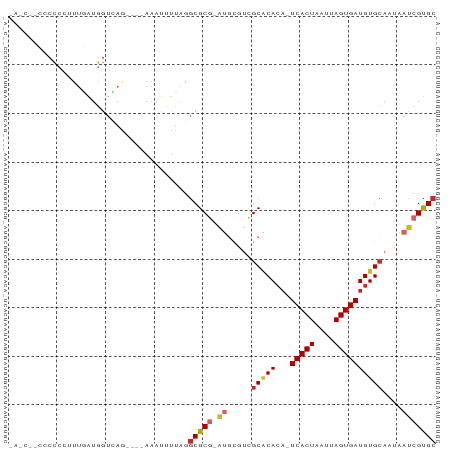
>dm3.chrX 7749412 81 - 22422827 CACC-UCCCCCUUUUGGUGGUCAG----AAAUUUUAGGCGCG-AUGCGUCGCACACA-UCACUAAUUAGUGAUGUGCAAUAAUCGUGC ((((-..........)))).....----.........(((((-((..((.(((((..-(((((....)))))))))).)).))))))) ( -25.50, z-score = -2.60, R) >droAna3.scaffold_13117 4189649 82 + 5790199 CAUCCGCCCCAAUUUGAUGGUUAG----AAAUUUCAGGCGCG-AUGCGUCGCACACA-UCACUAAUUAGUGAUGUGCAAUAAUCGUGC (((((((((((......)))...(----(....)).)))).)-))).......((((-(((((....)))))))))............ ( -23.30, z-score = -1.63, R) >droEre2.scaffold_4690 16662256 80 - 18748788 CACC--UCCCCCUUUAAUGGUCAG----AAAUUUUAGGCGCG-AUGCGUCGCACACA-UCACUAAUUAGUGAUGUGCAAUAAUCGUGC ....--....((......))....----.........(((((-((..((.(((((..-(((((....)))))))))).)).))))))) ( -21.90, z-score = -2.29, R) >droYak2.chrX 8206986 82 + 21770863 CACCUUCCCCCUUUUGAUGGUCAG----AAAUUUUAGGCGCG-AUGCGUCGCACACA-UCACUAAUUAGUGAUGUGCAAUAAUCGUGC .(((.((........)).)))...----.........(((((-((..((.(((((..-(((((....)))))))))).)).))))))) ( -23.00, z-score = -2.19, R) >droSim1.chrX 6200750 76 - 17042790 ------UCCCCCUUUGGUGGUCAG----AAAUUUUAGGCGCG-AUGCGUCGCACACA-UCACUAAUUAGUGAUGUGCAAUAAUCGUGC ------.((((....)).))....----.........(((((-((..((.(((((..-(((((....)))))))))).)).))))))) ( -23.10, z-score = -2.01, R) >droSec1.super_50 37044 75 - 201529 -------CCCCCUUUGGUGGUCAG----AAAUUUUAGGCGCG-AUGCGUCGCACACA-UCACUAAUUAGUGAUGUGCAAUAAUCGUGC -------((((....)).))....----.........(((((-((..((.(((((..-(((((....)))))))))).)).))))))) ( -23.00, z-score = -1.97, R) >droWil1.scaffold_181096 2697353 64 + 12416693 ------------------AAUCUG----AAAUUU-AAGCGCA-AUACCACGCGCACAUUCACUAAUUAGUGAUGUGCAAUAAUCGUGC ------------------.....(----(.....-..(((..-......)))((((((.((((....)))))))))).....)).... ( -16.10, z-score = -2.18, R) >droVir3.scaffold_13042 2551560 87 - 5191987 GACCCGCAAUCUAUUCGUGCCGAAACGUAAAAUUUAGGCGCGCGCGCGGCGCUCACA-UCACUAAUUAGUGAUGUGCAAUAAUCGUGC ...............((((((...............)))))).((((((....((((-(((((....)))))))))......)))))) ( -27.26, z-score = -1.18, R) >droMoj3.scaffold_6328 3416053 76 + 4453435 ------CGCCCCCUCGAUGGGCAU---CAUGCAGUUGCCGCACGAUUAUUGCACA---UCACUAAUUAGUGAUGUGAAUUCCGCGCGC ------.((((.......))))..---...((....))(((.((...(((.((((---(((((....))))))))))))..)).))). ( -26.10, z-score = -2.17, R) >consensus _A_C__CCCCCCUUUGAUGGUCAG____AAAUUUUAGGCGCG_AUGCGUCGCACACA_UCACUAAUUAGUGAUGUGCAAUAAUCGUGC .....................................(((((........(((((...(((((....))))))))))......))))) (-12.96 = -13.33 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:31 2011