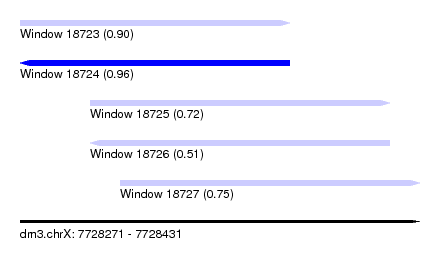
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,728,271 – 7,728,431 |
| Length | 160 |
| Max. P | 0.957468 |

| Location | 7,728,271 – 7,728,379 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Shannon entropy | 0.52028 |
| G+C content | 0.41001 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7728271 108 + 22422827 AAAGGGGGUCUUGGCUAAAAAUG---AGAAGAAAAAAAAAUAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA .(((..(((.((.((((......---.....................)))).)).))).((((((((((....)))))...))))).)))((((((.......))).))). ( -19.13, z-score = -0.07, R) >droSim1.chrX 6226135 106 + 17042790 GGAGGGGGUCUUGGCUAAAAAUG---AGAA--AAAAAAAAUAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA .(((..(((.((.((((......---....--...............)))).)).))).((((((((((....)))))...))))).)))((((((.......))).))). ( -19.71, z-score = -0.16, R) >droSec1.super_50 17586 103 + 201529 GGAGGGGGUCUUGGCUAAAAAUG---AGAA--AAAAA---UAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA .(((..(((.((.((((......---....--.....---.......)))).)).))).((((((((((....)))))...))))).)))((((((.......))).))). ( -19.85, z-score = -0.13, R) >droYak2.chrX 8178619 108 - 21770863 UGGGUGGGUUUUGGCACAAAAAA---AAAAAAAAAUGGAAAAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA ((((..(((((((.(........---...............(((.(((.......))).))).((((((....))))))..))))))))..))))................ ( -25.30, z-score = -2.04, R) >droEre2.scaffold_4690 16643692 111 + 18748788 UGGUGGGGUCUUGGCGAAAAAAUGACAAAAAAAAAAGAAGGGGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA .((..(((((..(((((......(((((............................)))))..))))).)))...((....))....))..))((((.......))))... ( -22.49, z-score = -0.56, R) >droAna3.scaffold_13117 4171969 83 - 5790199 ----------------------------GGAAGGGGUGGUGGAAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA ----------------------------..(((((((((..((..(((.......))).))..))))))......((....))....)))((((((.......))).))). ( -18.90, z-score = -0.62, R) >droWil1.scaffold_181096 2671249 99 - 12416693 -----GGGU---GGGAUGGAAGU----GAAGAGAAGAAGUAAAAAAAUUGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA -----(((.---.((....((((----(((((.((...((((.....)))).....)).))).))))))......((....))....))..)))................. ( -26.30, z-score = -3.13, R) >droMoj3.scaffold_6328 3383455 96 - 4453435 ---------------AGAUUGGUUCUGGGGUUGGUCCGAAAGAAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA ---------------.....(((((((..((((...(....).....)))).........((..(((((....)))))..)))))))))(((((((.......))).)))) ( -23.90, z-score = -0.73, R) >droGri2.scaffold_15203 5361138 84 + 11997470 ---------------------------UGUCAGCAGAAAAAAAAAAAAAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA ---------------------------..((................((((((..........))))))......((....)).))...(((((((.......))).)))) ( -16.40, z-score = -0.63, R) >consensus _____GGGU_UUGGCAAAAAAUG___AGAAAAAAAAAAAAAAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCA ...........................................................((((((((((....)))))...)))))...(((((((.......))).)))) (-16.30 = -16.30 + 0.00)

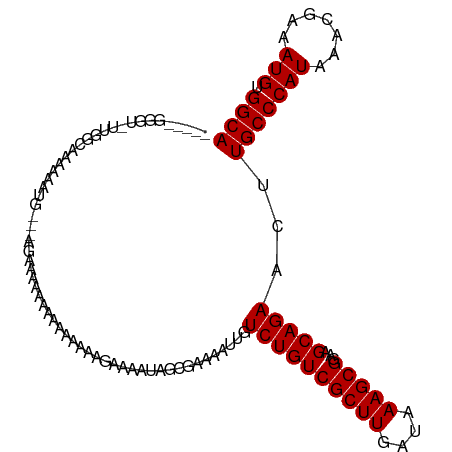
| Location | 7,728,271 – 7,728,379 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.07 |
| Shannon entropy | 0.52028 |
| G+C content | 0.41001 |
| Mean single sequence MFE | -15.44 |
| Consensus MFE | -13.60 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7728271 108 - 22422827 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUAUUUUUUUUUCUUCU---CAUUUUUAGCCAAGACCCCCUUU ((((.(((.......))))))).((.(((....(((((....))))).)))))..((((.((((.....................---......)))).))))........ ( -15.93, z-score = -1.05, R) >droSim1.chrX 6226135 106 - 17042790 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUAUUUUUUUU--UUCU---CAUUUUUAGCCAAGACCCCCUCC ((((.(((.......))))))).((.(((....(((((....))))).)))))..((((.((((...............--....---......)))).))))........ ( -16.01, z-score = -1.19, R) >droSec1.super_50 17586 103 - 201529 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUA---UUUUU--UUCU---CAUUUUUAGCCAAGACCCCCUCC ((((.(((.......))))))).((.(((....(((((....))))).)))))..((((.((((.......---.....--....---......)))).))))........ ( -16.15, z-score = -1.17, R) >droYak2.chrX 8178619 108 + 21770863 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUUUUCCAUUUUUUUUU---UUUUUUGUGCCAAAACCCACCCA ((.((((........(((((.(((....((...(((((....)))))...((......))))......))).)))))........---.....)))).))........... ( -13.90, z-score = -0.22, R) >droEre2.scaffold_4690 16643692 111 - 18748788 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCCCCUUCUUUUUUUUUUGUCAUUUUUUCGCCAAGACCCCACCA ((((.(((.......))))))).((..(((...(((((....)))))...(((((............................)))))........).))..))....... ( -16.39, z-score = -1.14, R) >droAna3.scaffold_13117 4171969 83 + 5790199 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUUCCACCACCCCUUCC---------------------------- ((((.(((.......))))))).((.(((....(((((....))))).)))))..............................---------------------------- ( -13.60, z-score = -1.41, R) >droWil1.scaffold_181096 2671249 99 + 12416693 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCAAUUUUUUUACUUCUUCUCUUC----ACUUCCAUCCC---ACCC----- ((((.(((.......))))))).((.(((....(((((....))))).)))))...............................----...........---....----- ( -13.60, z-score = -1.42, R) >droMoj3.scaffold_6328 3383455 96 + 4453435 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUUCUUUCGGACCAACCCCAGAACCAAUCU--------------- ((((.(((.......))))))).((((((..((((........(((((..........)))))..........)))).......))))))......--------------- ( -19.07, z-score = -1.53, R) >droGri2.scaffold_15203 5361138 84 - 11997470 UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUUUUUUUUUUUUUCUGCUGACA--------------------------- ((((.(((.......))))))).(((..((............((((((..........)))))).............)).))).--------------------------- ( -14.31, z-score = -0.39, R) >consensus UGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUUUUUUUUUUUUUUCU___AAUUUUUAGCCAA_ACCC_____ ((((.(((.......))))))).((.(((....(((((....))))).))))).......................................................... (-13.60 = -13.60 + -0.00)

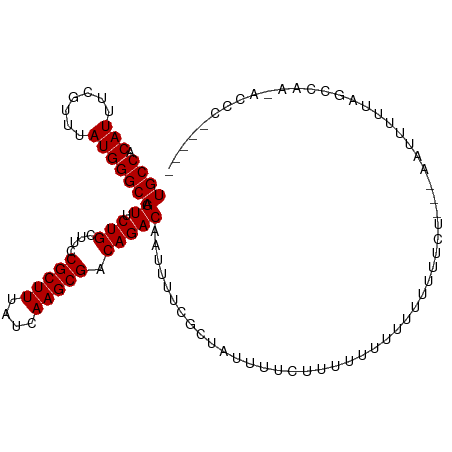
| Location | 7,728,299 – 7,728,419 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.23 |
| Shannon entropy | 0.18392 |
| G+C content | 0.44471 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.21 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
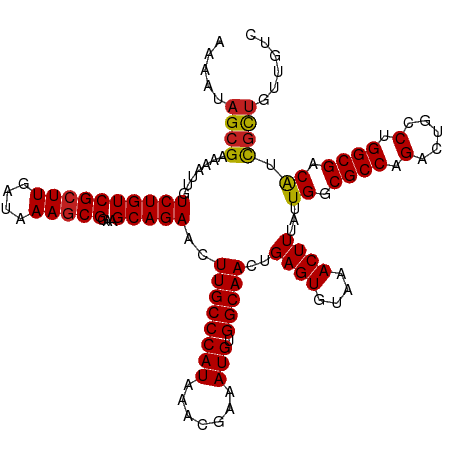
>dm3.chrX 7728299 120 + 22422827 AAAAAAAAAUAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC ..................((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -2.03, R) >droSim1.chrX 6226161 120 + 17042790 AAAAAAAAAUAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC ..................((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -2.03, R) >droSec1.super_50 17611 118 + 201529 --AAAAAAAUAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC --................((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -2.03, R) >droYak2.chrX 8178647 120 - 21770863 AAAAUGGAAAAGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC ..................((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -1.59, R) >droEre2.scaffold_4690 16643723 120 + 18748788 AAAAAGAAGGGGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC ..................((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -0.95, R) >droAna3.scaffold_13117 4171987 105 - 5790199 ---------------AUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC ---------------...((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -1.90, R) >dp4.chrXL_group1e 153688 106 - 12523060 --------------AAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC --------------....((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -1.90, R) >droPer1.super_14 1233305 106 + 2168203 --------------AAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC --------------....((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((((.....)))))... ( -30.40, z-score = -1.90, R) >droWil1.scaffold_181096 2671270 118 - 12416693 --AAGAAGUAAAAAAAUUGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGGCUGGCGAC --..(((((.......(((((.......((((((((((....)))))...)))))..((((((((.......))).))))).....)))))))))).....((((((.....)))))).. ( -31.01, z-score = -1.55, R) >droVir3.scaffold_13042 2519659 110 + 5191987 ----------GAAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAACGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCCGACCGACUGGCGAC ----------...............(((((.((((.(((.......((..((((...))))))...........(((.(((..((((....))))..))).))).))).)))).))))). ( -23.90, z-score = -0.58, R) >droMoj3.scaffold_6328 3383481 110 - 4453435 ----------GAAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCCGACCGACUGGCGAC ----------...............(((((.((((.(((.....((....))......................(((.(((..((((....))))..))).))).))).)))).))))). ( -26.40, z-score = -0.96, R) >droGri2.scaffold_15203 5361152 110 + 11997470 ----------AAAAAAAAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCCGACCGACUGGCGAC ----------...............(((((.((((.(((.....((....))......................(((.(((..((((....))))..))).))).))).)))).))))). ( -26.40, z-score = -0.94, R) >consensus __________AGAAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGAC ..................((.((.....((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..)).))(((.(.....).)))... (-25.12 = -25.21 + 0.08)
| Location | 7,728,299 – 7,728,419 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.23 |
| Shannon entropy | 0.18392 |
| G+C content | 0.44471 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -21.93 |
| Energy contribution | -21.86 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
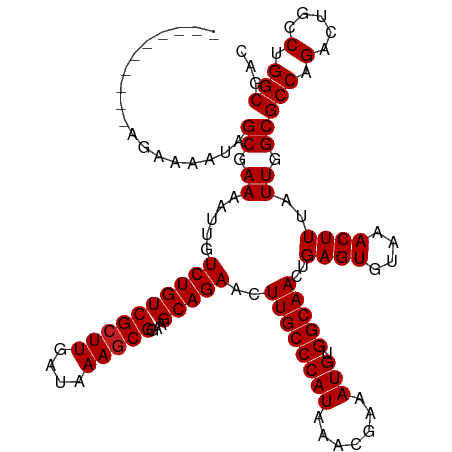
>dm3.chrX 7728299 120 - 22422827 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUAUUUUUUUUU (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).)))))........................... ( -27.60, z-score = -1.88, R) >droSim1.chrX 6226161 120 - 17042790 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUAUUUUUUUUU (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).)))))........................... ( -27.60, z-score = -1.88, R) >droSec1.super_50 17611 118 - 201529 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUAUUUUUUU-- (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).))))).........................-- ( -27.60, z-score = -1.92, R) >droYak2.chrX 8178647 120 + 21770863 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCUUUUCCAUUUU (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).)))))........................... ( -27.60, z-score = -1.85, R) >droEre2.scaffold_4690 16643723 120 - 18748788 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCCCCUUCUUUUU (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).)))))........................... ( -27.60, z-score = -1.64, R) >droAna3.scaffold_13117 4171987 105 + 5790199 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAU--------------- (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).)))))............--------------- ( -27.60, z-score = -1.76, R) >dp4.chrXL_group1e 153688 106 + 12523060 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUU-------------- (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).))))).............-------------- ( -27.60, z-score = -1.77, R) >droPer1.super_14 1233305 106 - 2168203 GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUU-------------- (((....))).(((((.(((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))).))))).............-------------- ( -27.60, z-score = -1.77, R) >droWil1.scaffold_181096 2671270 118 + 12416693 GUCGCCAGCCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCAAUUUUUUUACUUCUU-- ((((((((.....)))))((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...))))).............................-- ( -26.10, z-score = -1.65, R) >droVir3.scaffold_13042 2519659 110 - 5191987 GUCGCCAGUCGGUCGGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGUUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUUC---------- (((((........((((.((......(((....)))...(((((.(((.......)))))))).....)).)))).........))))).....................---------- ( -21.43, z-score = -0.06, R) >droMoj3.scaffold_6328 3383481 110 + 4453435 GUCGCCAGUCGGUCGGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUUC---------- (((((.....(((.....))).(((((((.......((((((((.(((.......))))))))...))).....)))))))...))))).....................---------- ( -22.90, z-score = -0.28, R) >droGri2.scaffold_15203 5361152 110 - 11997470 GUCGCCAGUCGGUCGGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUUUUUUUU---------- (((((.....(((.....))).(((((((.......((((((((.(((.......))))))))...))).....)))))))...))))).....................---------- ( -22.90, z-score = -0.15, R) >consensus GUCGCCAGGCAGUCUGGCGCCAAUAAAGUUUACACUCAGUUGCCACAUUUCGUUUAUGGGCAAGUUCUGCUUCCGCUUUAUCAAGCGACAGACAAUUUUCGCUAUUUUCU__________ ((((((.........)))((..(((((((.......((((((((.(((.......))))))))...))).....)))))))...)))))............................... (-21.93 = -21.86 + -0.08)

| Location | 7,728,311 – 7,728,431 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Shannon entropy | 0.11643 |
| G+C content | 0.46017 |
| Mean single sequence MFE | -36.11 |
| Consensus MFE | -27.80 |
| Energy contribution | -27.76 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
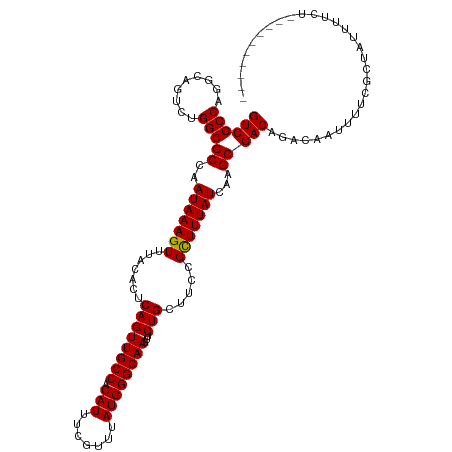
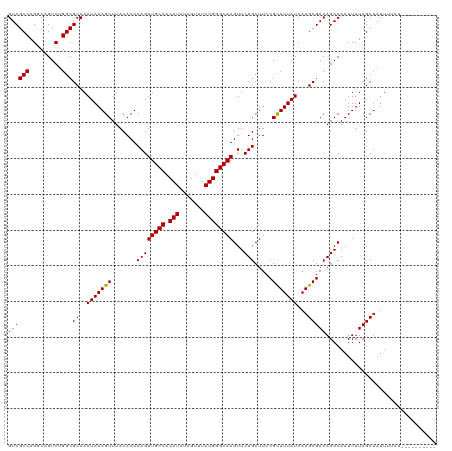
>dm3.chrX 7728311 120 + 22422827 AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAUCGCUGUUGUC ..((((((((......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((((.....)))))).))))))))))... ( -39.30, z-score = -3.04, R) >droSim1.chrX 6226173 120 + 17042790 AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAUCGCUGUUGUC ..((((((((......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((((.....)))))).))))))))))... ( -39.30, z-score = -3.04, R) >droSec1.super_50 17621 120 + 201529 AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAUCGCUGUUGUC ..((((((((......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((((.....)))))).))))))))))... ( -39.30, z-score = -3.04, R) >droYak2.chrX 8178659 120 - 21770863 AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAUCGCUGUUGUC ..((((((((......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((((.....)))))).))))))))))... ( -39.30, z-score = -3.04, R) >droEre2.scaffold_4690 16643735 120 + 18748788 AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAUCGCUGUUGUC ..((((((((......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((((.....)))))).))))))))))... ( -39.30, z-score = -3.04, R) >droAna3.scaffold_13117 4171987 117 - 5790199 ---AUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAUCGCUGUUGUC ---(((((((......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((((.....)))))).))))))))).... ( -37.50, z-score = -2.44, R) >dp4.chrXL_group1e 153688 118 - 12523060 --AAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAACGCUGUUGUC --(((((((.......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..(((.((((((.....)))))).))))))))))... ( -38.50, z-score = -2.86, R) >droPer1.super_14 1233305 118 + 2168203 --AAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAACGCUGUUGUC --(((((((.......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))..(((.((((((.....)))))).))))))))))... ( -38.50, z-score = -2.86, R) >droWil1.scaffold_181096 2671280 120 - 12416693 AAAAUUGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGGCUGGCGACAUUGCUGUUGUC ..(((.((((......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((((.....)))))).)))))).)))... ( -32.80, z-score = -0.97, R) >droVir3.scaffold_13042 2519661 114 + 5191987 AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAACGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCCGACCGACUGGCGACAACGUU------ ...............((.(((((((........(((..((.....((((((((.......))).)))))..((((....)))).....))..))).......)))))))))...------ ( -25.96, z-score = -0.45, R) >droMoj3.scaffold_6328 3383483 114 - 4453435 AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCCGACCGACUGGCGACAACAUU------ ..............(((.(((((((........(((..((.....((((((((.......))).)))))..((((....)))).....))..))).......))))))))))..------ ( -27.76, z-score = -0.94, R) >droGri2.scaffold_15203 5361154 120 + 11997470 AAAAAAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCCGACCGACUGGCGACGUCGCUGUCGCU .....(((((.....(((.(((((((....))))((..((.....((((((((.......))).)))))..((((....)))).....))..))))).))).((((....)))).))))) ( -35.80, z-score = -1.63, R) >consensus AAAAUAGCGAAAAUUGUCUGUCGCUUGAUAAAGCGGAAGCAGAACUUGCCCAUAAACGAAAUGUGGCAACUGAGUGUAAACUUUAUUGGCGCCAGACUGCCUGGCGACAUCGCUGUUGUC .....((((.......((((((((((....)))))...)))))..((((((((.......))).)))))..((((....))))...((.((((.(.....).)))).)).))))...... (-27.80 = -27.76 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:26 2011