
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,555,836 – 10,555,948 |
| Length | 112 |
| Max. P | 0.716113 |

| Location | 10,555,836 – 10,555,948 |
|---|---|
| Length | 112 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.81 |
| Shannon entropy | 0.58659 |
| G+C content | 0.44497 |
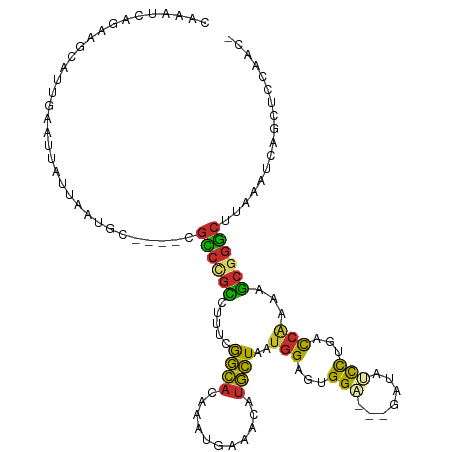
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -12.86 |
| Energy contribution | -11.72 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.81 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
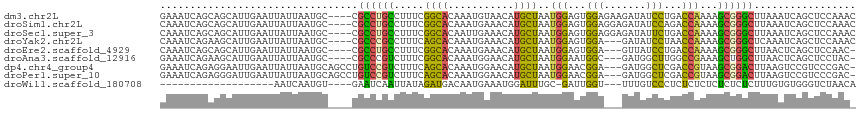
>dm3.chr2L 10555836 112 - 23011544 GAAAUCAGCAGCAUUGAAUUAUUAAUGC----CGCCUGCCUUUCGGCACAAAUGUAACAUGCUAAUGGAGUGGAGAAGAUAUCCUGACCAAAAGCGGGCUUAAAUCAGCUCCAAAC ......(((((((((((....)))))))----....((((....))))...........))))..((((((.((.(((....((((........)))))))...)).))))))... ( -28.60, z-score = -1.26, R) >droSim1.chr2L 10359749 112 - 22036055 CAAAUCAGCAGCAUUGAAUUAUUAAUGC----CGCCUGCCUUUCGGCACAAAUGAAACAUGCUAAUGGAGUGGAGGAGAUAUCCAGACCAAAAGCGGGCUUAAAUCAGCUCCAAAC ....(((((.(((((((....)))))))----.)).((((....))))....))).....(((..(((..((((.......))))..)))..)))(((((......))).)).... ( -30.70, z-score = -1.76, R) >droSec1.super_3 5982103 112 - 7220098 CAAAUCAGCAGCAUUGAAUUAUUAAUGC----CGCCUGCCUUUCGGCACAAUUGAAACAUGCUAAUGGAGUGGAGGAGAUAUUCUGACCAAAAGCGGGCUUAAAUCAGCUCCAAAC ....(((((.(((((((....)))))))----.)).((((....))))....))).....(((..(((..(((((......))))).)))..)))(((((......))).)).... ( -26.90, z-score = -0.45, R) >droYak2.chr2L 6965058 109 - 22324452 CAAAUCAGAAGCAUUGAAUUAUUAAUGC----CGCCCGCCUUUCAGCACAAAUGAAACAUGCUAAUGGAGUGGA---GAUAUCCUAACCAAAAGCGGGCUCAAAUCAGCUCCAAAC .......((.(((((((....)))))))----.((((((.(((((.......)))))........(((...(((---....)))...)))...)))))).....)).......... ( -27.00, z-score = -2.12, R) >droEre2.scaffold_4929 11769493 108 + 26641161 CAAAUCAGCAGCAUUGAAUUAUUAAUGC----CGCCUGCCUUUCGGCACAAAUGAAACAUGCUAAUGGAGUGGA---GUUAUCCUGACCAAAAGCGGGCUUAACUCAGCUCCAAC- ....(((((.(((((((....)))))))----.)).((((....))))....)))..........((((((.((---((((.((((........))))..)))))).))))))..- ( -35.50, z-score = -3.51, R) >droAna3.scaffold_12916 14405611 108 - 16180835 GAAAUCAGAAGCAUUGAAUUAUUAAUGC----CGCCCGUCUUUCGGCACAAAUGGAACAUGCUAAUGGAAUGGC---GAUGGCUUGGCCGAAAGCUGGCUUAACUCAGCUCCUAC- .......((.(((((((....)))))))----.(((.(.((((((((.(((......(((((((......))))---.)))..)))))))))))).))).....)).........- ( -29.90, z-score = -1.00, R) >dp4.chr4_group4 2763320 112 - 6586962 GAAAUCAGAGGAAUUGAAUUAUUAAUGCAGCCUGUCCGUCUUUCAGCACAAAUGGAACAUGCUAAUGGAACGGA---GAUGGCUCGACCGUAAGCGGACUUAAGUCCGUCCCGAC- .........((.......((((...((.(((((.(((((((...((((...........))))...)).)))))---.).))))))...))))((((((....)))))).))...- ( -28.50, z-score = -0.61, R) >droPer1.super_10 1771536 112 - 3432795 GAAAUCAGAGGGAUUGAAUUAUUAAUGCAGCCUGUCCGUCUUUCAGCACAAAUGGAACAUGCUAAUGGAACGGA---GAUGGCUCGACCGUAAGCGGACUUAAGUCCGUCCCGAC- .........(((......((((...((.(((((.(((((((...((((...........))))...)).)))))---.).))))))...))))((((((....)))))))))...- ( -31.60, z-score = -1.19, R) >droWil1.scaffold_180708 1766300 89 + 12563649 -------------------AAUCAAUGU----GAAUCAAUUAUAGAUGACAAUGAAAUGGAUUUGC-GAUUGGU---UUUGUCCCUCUCUCUCUCUCUCUUUGUGUGGGUCUAACA -------------------((((((((.----.((((.(((.((........)).))).))))..)-.))))))---)..(.(((.(.(.............).).))).)..... ( -14.12, z-score = 0.57, R) >consensus CAAAUCAGAAGCAUUGAAUUAUUAAUGC____CGCCCGCCUUUCGGCACAAAUGAAACAUGCUAAUGGAGUGGA___GAUAUCCUGACCAAAAGCGGGCUUAAAUCAGCUCCAAC_ .................................((((((.....((((...........))))..(((...(((.......)))...)))...))))))................. (-12.86 = -11.72 + -1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:28 2011