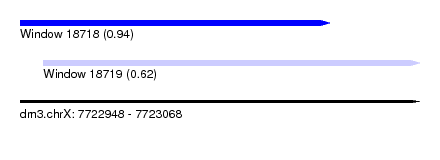
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,722,948 – 7,723,068 |
| Length | 120 |
| Max. P | 0.940502 |

| Location | 7,722,948 – 7,723,041 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.74 |
| Shannon entropy | 0.27344 |
| G+C content | 0.37702 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -19.00 |
| Energy contribution | -18.90 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940502 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
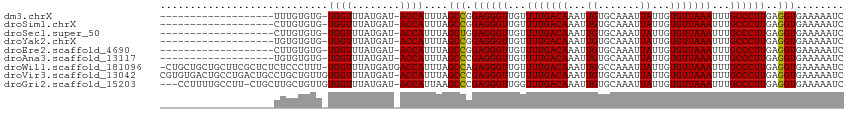
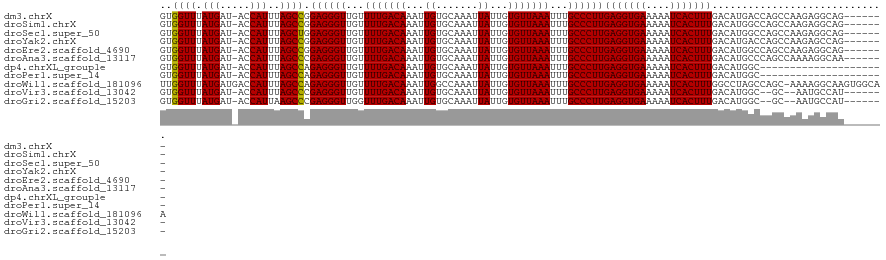
>dm3.chrX 7722948 93 + 22422827 -------------------UUUGUGUG-UGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC -------------------.......(-((((.......-)))))...(((.((((((...(((((((....((....)).....)))))))...))))))..)))........ ( -22.00, z-score = -1.79, R) >droSim1.chrX 6168925 93 + 17042790 -------------------CUUGUGUG-UGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC -------------------.......(-((((.......-)))))...(((.((((((...(((((((....((....)).....)))))))...))))))..)))........ ( -22.00, z-score = -1.65, R) >droSec1.super_50 12398 93 + 201529 -------------------CUUGUGUG-UGGUUUAUGAU-ACCAUUUAGCUGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC -------------------........-((((.......-))))((((.((.((((((...(((((((....((....)).....)))))))...)))))).)).))))..... ( -23.70, z-score = -2.62, R) >droYak2.chrX 8173167 93 - 21770863 -------------------UGUGUGUG-UGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC -------------------.......(-((((.......-)))))...(((.((((((...(((((((....((....)).....)))))))...))))))..)))........ ( -22.00, z-score = -1.62, R) >droEre2.scaffold_4690 16638743 93 + 18748788 -------------------CUUGUGUG-UGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC -------------------.......(-((((.......-)))))...(((.((((((...(((((((....((....)).....)))))))...))))))..)))........ ( -22.00, z-score = -1.65, R) >droAna3.scaffold_13117 4167441 93 - 5790199 -------------------UGUGUGUG-UGGUUUAUGAU-ACCAUUUAGCCCGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC -------------------.......(-((((.......-)))))...((((((((((...(((((((....((....)).....)))))))...))))))).)))........ ( -24.50, z-score = -2.74, R) >droWil1.scaffold_181096 2665762 112 - 12416693 -CUGCUGCUGCUUCGCUCUCUCCCUUU-UGGUUUAUGAUGACCAUUUAGCCAGAGGGUUGUUUUGACAAAUUGGCCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC -..((....))(((((.(((.......-(((((.(((.....)))..)))))((((((...(((((((...(((.....)))...)))))))...))))))))))))))..... ( -28.40, z-score = -1.47, R) >droVir3.scaffold_13042 2503643 113 + 5191987 CGUGUGACUGCCUGACUGCCUGCUGUUGUGGUUUAUGAU-ACCAUUUAGCCCGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC .................((((((((..(((((.......-))))).)))).(((((((...(((((((....((....)).....)))))))...)))))))))))........ ( -29.20, z-score = -2.30, R) >droGri2.scaffold_15203 5354117 109 + 11997470 ---CCUUUUGCCUU-CUGCUUGCUGUUGUGGUUUAUGAU-ACCAUUAAGCCCGAGGGUUGGUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC ---..(((..(((.-......(((...(((((.......-)))))..))).(((((((..((((((((....((....)).....))))))))..))))))))))..))).... ( -34.10, z-score = -4.09, R) >consensus ___________________CUUGUGUG_UGGUUUAUGAU_ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUC ............................((((........))))....(((.((((((...(((((((...((.......))...)))))))...))))))..)))........ (-19.00 = -18.90 + -0.10)
| Location | 7,722,955 – 7,723,068 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
| Shannon entropy | 0.19951 |
| G+C content | 0.39139 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -23.54 |
| Energy contribution | -23.45 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.616549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
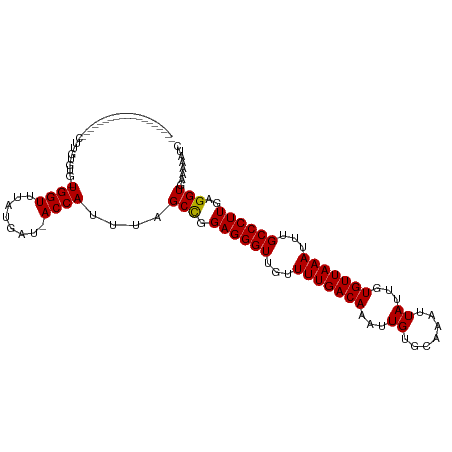
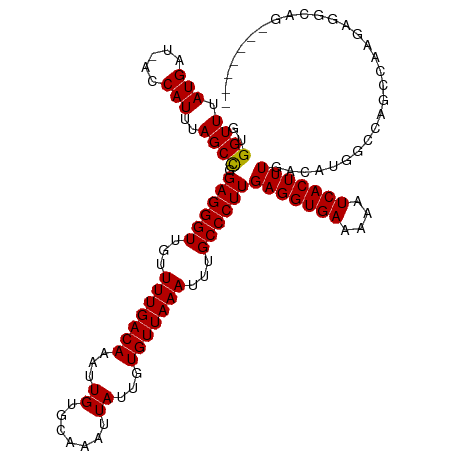
>dm3.chrX 7722955 113 + 22422827 GUGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGACCAGCCAAGAGGCAG------- .(((((.(((..-............((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))..))))))))(((....)))..------- ( -31.30, z-score = -1.82, R) >droSim1.chrX 6168932 113 + 17042790 GUGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAG------- (((((.......-)))))...((((((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))....))))..(((....)))..------- ( -33.10, z-score = -1.89, R) >droSec1.super_50 12405 113 + 201529 GUGGUUUAUGAU-ACCAUUUAGCUGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAG------- .(((((.(((..-.(((......))).((((...(((((((....((....)).....)))))))...))))(..((((((....))))))..)))))))))(((....)))..------- ( -31.00, z-score = -1.26, R) >droYak2.chrX 8173174 113 - 21770863 GUGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGACCAGCCAAGAGCCAG------- .(((((((((..-..)))...((.((.((((...(((((((....((....)).....)))))))...))))(..((((((....))))))..).....)).))...)))))).------- ( -28.10, z-score = -1.09, R) >droEre2.scaffold_4690 16638750 113 + 18748788 GUGGUUUAUGAU-ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAG------- (((((.......-)))))...((((((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))....))))..(((....)))..------- ( -33.10, z-score = -1.89, R) >droAna3.scaffold_13117 4167448 113 - 5790199 GUGGUUUAUGAU-ACCAUUUAGCCCGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGCCCAGCCAAAAGGCAA------- ((((((((((..-..)))...((((((((((...(((((((....((....)).....)))))))...))))))).)))...))))))).............(((....)))..------- ( -30.30, z-score = -1.54, R) >dp4.chrXL_group1e 147774 99 - 12523060 GUGGUUUAUGAU-ACCAUUUAGCCAGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC--------------------- (((((.......-)))))...((((((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))....))))--------------------- ( -28.40, z-score = -2.62, R) >droPer1.super_14 1227501 99 + 2168203 GUGGUUUAUGAU-ACCAUUUAGCCAGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC--------------------- (((((.......-)))))...((((((((((...(((((((....((....)).....)))))))...))))))(((((((....)))))))....))))--------------------- ( -28.40, z-score = -2.62, R) >droWil1.scaffold_181096 2665787 120 - 12416693 UUGGUUUAUGAUGACCAUUUAGCCAGAGGGUUGUUUUGACAAAUUGGCCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGGCCUAGCCAGC-AAAAGGCAAGUGGCAA .(((((......)))))....(((.((((((...(((((((...(((.....)))...)))))))...))))))(((((((....))))))))))...((((((-.....))...)))).. ( -35.20, z-score = -1.26, R) >droVir3.scaffold_13042 2503670 109 + 5191987 GUGGUUUAUGAU-ACCAUUUAGCCCGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC--GC--AAUGCCAU------- ((((((((((..-..)))...((((((((((...(((((((....((....)).....)))))))...))))))).)))...)))))))......(((((--..--...)))))------- ( -31.00, z-score = -1.99, R) >droGri2.scaffold_15203 5354140 109 + 11997470 GUGGUUUAUGAU-ACCAUUAAGCCCGAGGGUUGGUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGC--GC--AAUGCCAU------- ((((((((((..-..)))...((((((((((..((((((((....((....)).....))))))))..))))))).)))...)))))))......(((((--..--...)))))------- ( -33.50, z-score = -2.42, R) >consensus GUGGUUUAUGAU_ACCAUUUAGCCGGAGGGUUGUUUUGACAAAUUGUGCAAAUUAUUGUGUUAAAUUUGCCCUUGAGGUGAAAAAUCACUUUGACAUGGCCAGCCAAGAGGCAG_______ ..((((.(((.....)))..)))).((((((...(((((((...((.......))...)))))))...))))))(((((((....)))))))............................. (-23.54 = -23.45 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:20 2011