| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,652,048 – 7,652,144 |
| Length | 96 |
| Max. P | 0.949538 |

| Location | 7,652,048 – 7,652,144 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.34 |
| Shannon entropy | 0.43772 |
| G+C content | 0.47744 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949538 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
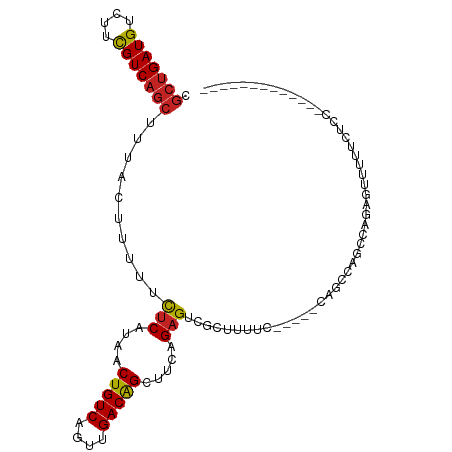
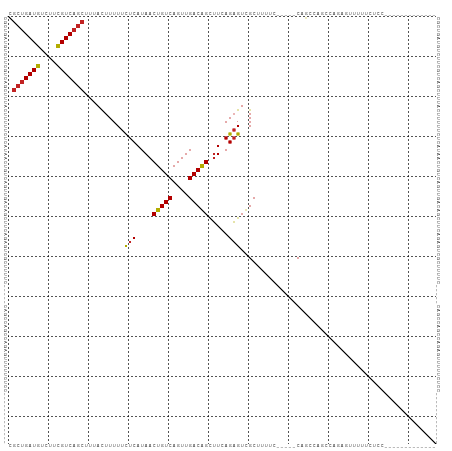
>dm3.chrX 7652048 96 + 22422827 CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACGGCUUCAGAGUCGCUUUUC-----CAGCCAGCCAGAGUUUUUCUCCUCUGUCC------ .(((((((....)))))))...................(((....)))((((...(((......)))-----.))))...(((((........)))))...------ ( -23.80, z-score = -2.00, R) >droSim1.chrX 6122981 88 + 17042790 CGGGGAUGUGUUUGUCAG-UUUAUUUUUUUUCAUAACUGUCAGUUGACGGUUUCAGAGUCGCUUUUC-----CAGCCAGCCAGAGUUUUUCUCC------------- .(((((....((((.(..-..........(((..(((((((....)))))))...)))..(((....-----.)))..).)))).....)))))------------- ( -16.40, z-score = -0.20, R) >droSec1.super_24 857045 89 + 912625 CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACGGCUUCAGAGUCGCUUUUC-----CAGCCAGCCAGAGUUUUUCUCC------------- .(((((((....)))))))..........(((......(((....)))((((...(((......)))-----.)))).....))).........------------- ( -21.30, z-score = -1.65, R) >droYak2.chrX 8105707 97 - 21770863 CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACGGCUUCAGAGUCGCUUUUCCUUUCCCGCCAGCCAGAGUUUUUCCACUCC---------- .(((((((....))))))).......................((((.(((.....(((......))).....))).))))..((((......)))).---------- ( -20.30, z-score = -1.59, R) >droEre2.scaffold_4690 16577316 92 + 18748788 CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACGGCUUCAGAGUCGCUUUUC-----CAGCCAGCCAGAGUUUUUCUCGUCC---------- .(((((((....)))))))..........................(((((((...(((......)))-----.)))).....(((.....)))))).---------- ( -21.40, z-score = -1.36, R) >droAna3.scaffold_13117 4106784 89 - 5790199 CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUCGCUUUUC-----CAGCCAGUCGGAGUUUUCCUCC------------- .(((((((....))))))).................(((((....))))).....((.(.(((....-----.))).).))((((.....))))------------- ( -23.20, z-score = -2.48, R) >dp4.chrXL_group1e 11678226 103 + 12523060 CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCUUUUUC----CAUCCAGCCAGAGCUGUCCUCCGUUUCCGCCUCGU .(((((((....)))))))..........................((((((((..(.((((.......----....))))).))))))))................. ( -22.50, z-score = -1.51, R) >droPer1.super_14 1140672 103 + 2168203 CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCUUUUUC----CAUCCAGCCAGAGUUGUCCUCCGUUUCCGCCUCGU .(((((((....)))))))..........................((((((((..(.((((.......----....))))).))))))))................. ( -20.10, z-score = -1.02, R) >droMoj3.scaffold_6328 1202346 80 + 4453435 GGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCGUCAGUGUUGCCACCC-----CGCGCAACGAACG---------------------- ((((((((....))))))))................(((((....)))))(((...(((((((....-----.).)))))).)))---------------------- ( -23.50, z-score = -2.24, R) >droVir3.scaffold_13042 2814481 75 - 5191987 GGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCCACCC-----CGCCAAAC--------------------------- ((((((((....)))))))).........(((....(((((....))))).....))).........-----........--------------------------- ( -17.70, z-score = -1.66, R) >droGri2.scaffold_15203 6801129 77 - 11997470 GGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUUGCCACCC-----CGCCAAACGA------------------------- ((((((((....)))))))).........(((....(((((....))))).....))).........-----..........------------------------- ( -17.70, z-score = -1.22, R) >consensus CGCUGAUGUCUUCGUCAGCUUUACUUUUUCUCAUAACUGUCAGUUGACAGCUUCAGAGUCGCUUUUC_____CAGCCAGCCAGAGUUUUUCUCC_____________ .(((((((....)))))))..........(((....(((((....))))).....)))................................................. (-15.50 = -15.45 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:12 2011