| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,646,811 – 7,646,906 |
| Length | 95 |
| Max. P | 0.584877 |

| Location | 7,646,811 – 7,646,906 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 60.89 |
| Shannon entropy | 0.69470 |
| G+C content | 0.57663 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7646811 95 + 22422827 AAGUGAUUUAUGCUGAGCACCUGG-CUUAGCU-CGAAUCAGCA----CCCCCACCACAGC-------------CCCUUGCCCUACAGCGCAUCGCAACCCAUGGCCCUGCCACG ...((((((..(((((((.....)-)))))).-.))))))(((----...(((.....((-------------....(((.(....).)))..))......)))...))).... ( -23.10, z-score = -1.13, R) >droAna3.scaffold_13117 4103090 86 - 5790199 AAGUGAUUUAUGCUGAGCACCUGG-CUUAGCU-CGGAUCCGCCC---CUCCGGCGCCGCCU------------CCCCUAUCAACUGGUCCUCUUCCGGCCACC----------- .((((((....(((((((.....)-)))))).-.((...((((.---....))))...)).------------.....))).)))((((.......))))...----------- ( -24.40, z-score = -0.76, R) >droEre2.scaffold_4690 16572540 112 + 18748788 AAGUGAUUUAUGCCGAGCACCUGG-CUUAGCU-CGAUUCAGCCCCUUCCCCCUCCACACCUGACCCCACCAUACCGCCGCCCCAUCGUGCUGCUCAGCCCCUAGCCCUGCCACG ..(((......((.(.((....((-((.(((.-....((((..................))))............((((......)).)).))).))))....)).).))))). ( -17.07, z-score = 0.16, R) >droSec1.super_24 852020 95 + 912625 AAGUGAUUUAUGCUGAGCACCUGG-CUUAGCU-CGAAUCAGU-----CCCCCACCUCACCC------------CUCCUGCCGUAUCGCGCCCCGCAAUUCCUGGCCCUACCACG ...((((((..(((((((.....)-)))))).-.))))))..-----..............------------.....((((.((.(((...))).))...))))......... ( -21.80, z-score = -2.04, R) >droSim1.chrX 6117733 95 + 17042790 AAGUGAUUUAUGCUGAGCACCUGG-CUUAGCU-CGAAUCAGU-----CCCCAACCUCACCG------------CCCCUGCCGUAUCGCGCCCCGCAAUUCCUGGCCCUUCCACG ...((((((..(((((((.....)-)))))).-.))))))..-----..............------------.....((((.((.(((...))).))...))))......... ( -21.80, z-score = -1.52, R) >droWil1.scaffold_181096 2574300 98 - 12416693 AAGUGAUUUAUGUUAAGCACCUGGACUUAACUUCCAGUCCUUUUAGAGCUCUGUCUAGGAU-----------AUUGGGUAACCAUUCUGAUCCUUUGUCCUUUAUGCUG----- ...............((((...(((((........)))))...((((......))))((((-----------(..((((..........))))..)))))....)))).----- ( -19.90, z-score = -0.22, R) >consensus AAGUGAUUUAUGCUGAGCACCUGG_CUUAGCU_CGAAUCAGC_____CCCCCACCUCACCU____________CCCCUGCCCUAUCGCGCUCCGCAAUCCCUGGCCCUGCCACG ....(((((..(((((((.....).))))))...)))))........................................................................... ( -8.68 = -8.52 + -0.16)

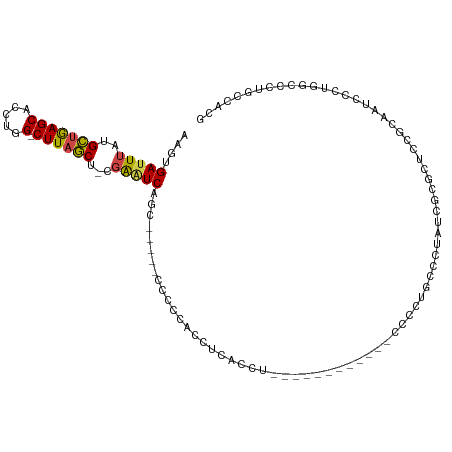

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:11 2011