
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,646,349 – 7,646,494 |
| Length | 145 |
| Max. P | 0.899167 |

| Location | 7,646,349 – 7,646,494 |
|---|---|
| Length | 145 |
| Sequences | 9 |
| Columns | 147 |
| Reading direction | reverse |
| Mean pairwise identity | 68.06 |
| Shannon entropy | 0.68174 |
| G+C content | 0.39520 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -13.61 |
| Energy contribution | -13.67 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.899167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
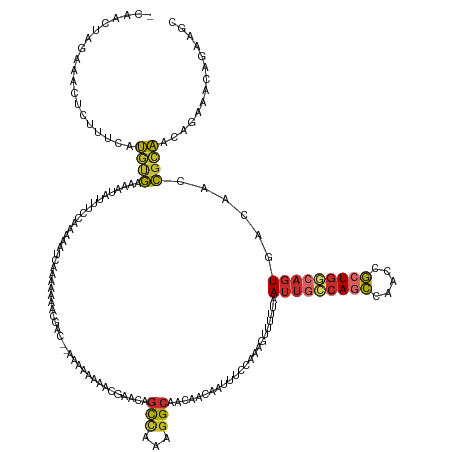
>dm3.chrX 7646349 145 - 22422827 GAAACUAGAAACUCUUUCAUGUGAAAAUAUUUCCAAAAAUCAGGAAAACGAC--AAAAAAAAAGGACAGCCAAAGGCAACAACAAUUUACGAAGUUUUUAAUUGCCAGCCAACCGCUGGGAGUGACAACCGCAACAGAAAUAGCAGC .....((((((((...((..(((((....(((((........))))).....--..............(((...)))........)))))))))))))))(((.(((((.....))))).))).......((..........))... ( -23.50, z-score = -0.13, R) >droSim1.chrX 6117273 144 - 17042790 -GAACUAGAAACUCUUUCAUGUGAAAAUAUUUCCAAAAAUCAGGAAAACGAC--AAAAAAAACGUACAGCCAAAGGCAACAACAAUUUCCAAAGUUUUUAAUUGCCAGCCAACCGCUGGCAGUGACAACCGCAACAGAAACAACAGC -....((((((((.((((....))))................((((((((..--........)))...(((...)))........)))))..))))))))(((((((((.....)))))))))........................ ( -28.00, z-score = -2.28, R) >droSec1.super_24 851558 146 - 912625 -GAACUAGAAACUCUUUCAUGUGAAAAUAUUUCCAAAAAUCAGGAAAACGACAAAAAAAAAACGAACAGCCAAAGGCAACAACAAUUUCCAAAGUUUUUAAUUGCCAGCCAACCGCUGGCAGUGACAACCGCAACAGAAACAGCAGC -....((((((((..(((.(((....)))(((((........)))))................)))..(((...)))...............))))))))(((((((((.....))))))))).......((..........))... ( -27.50, z-score = -1.85, R) >droYak2.chrX 8100229 141 + 21770863 -GAACUAGAAACUCUUUCAUGCGAAAAUAUUUCCAAAAAUCAGGAAAAA-----ACAAAAAACGUACAGCCAAAGGCAACAGCAUUUUCCAAAGUUUUUAAUUGCCAGCCAACCGCUGGCAGUGACAACCGCAACAGAAACAGAAAC -..........((.((((.(((((((((.(((((........)))))..-----..............(((...)))......))))))....(((....(((((((((.....)))))))))...))).)))...)))).)).... ( -30.60, z-score = -3.29, R) >droEre2.scaffold_4690 16572048 138 - 18748788 -CAACUAGAAACUCUUUCAUGUGAAAAUAUUUCACAACACAAAAAAAAU-----ACAAAAACU---CAGCCAAGGGCAACAACCAUUUCGAAAGUUUUUAAUUGCCAGCCAACCGAUGGCAGUGACAACCGCAACAGAAACAAAAGC -(((.((((((((..(((.((((((.....)))))).............-----.........---..(((...)))............))))))))))).)))...((((.....)))).(((.....)))............... ( -22.90, z-score = -1.50, R) >droAna3.scaffold_13117 4102635 124 + 5790199 ----------------GGGCGGGGAAUGCUGUUUGCCAA--AAAAAAAUA----GAAACAAGUUAUGGGCCAAAGGCAACAG-AAUUUGCAAAGUUUUUAAUUGCCAGCCGACCGCUGGCAGUGACAAGCGGGGCGGGAAUCGAAAC ----------------.((((.....)))).((((((..--.........----....((((((.((.(((...)))..)).-))))))....((((...(((((((((.....)))))))))...))))..))))))......... ( -33.30, z-score = -0.09, R) >dp4.chrXL_group1e 11668334 143 - 12523060 -CCCGUAGAAGCCAUUUCAUGUGAAAAUACGAAUGAAAA--AAAAAAGCGACAGAGCAAAAACAAACAGCUGAAGGCAACAA-AAUUUGCAAAGUUUUUAAUUGCCAGCCAACCGCUGUCAGUGACAACCGCAGCAGGAACAGAAGC -.((..........((((((.((......)).)))))).--......((((((((((...........)))...(((((.((-((((.....))))))...))))).........))))).(((.....))).)).))......... ( -25.10, z-score = 0.34, R) >droPer1.super_14 1131467 141 - 2168203 -CCCGUAGAAGCCAUUUCAUGUGAAAAUACGAAUG--AA--AAAAAAGCGACAGAGCAAAAACAAACAGCUGAAGGCAACAA-AAUUUGCAAAGUUUUUAAUUGCCAGCCAACCGCUGUCAGUGACAACCGCAGCAGGAACAGAAGC -.((..........((((((.((......)).)))--))--).....((((((((((...........)))...(((((.((-((((.....))))))...))))).........))))).(((.....))).)).))......... ( -25.10, z-score = 0.37, R) >droWil1.scaffold_181096 2573796 132 + 12416693 -------ACCUUCAACGAACUUUGCAUACCCUUAAAAUACCAAAAAAA------AAAAAAAAGGACCUAUAAUGAGCAUAAGAAGUAAUUGAAG--CUUAAGUAAAAAAAAAAGUAUGUAUAUGGCAAGAGAGACUUGAAAUUUGGC -------........((((...(((((((((((...............------......))))........(((((................)--)))).............))))))).....((((.....))))...)))).. ( -13.69, z-score = 0.81, R) >consensus _CAACUAGAAACUCUUUCAUGUGAAAAUAUUUCCAAAAAUCAAAAAAACGAC__AAAAAAAACGAACAGCCAAAGGCAACAACAAUUUCCAAAGUUUUUAAUUGCCAGCCAACCGCUGGCAGUGACAACCGCAACAGAAACAGAAGC ...................((((.............................................(((...))).......................(((((((((.....)))))))))......)))).............. (-13.61 = -13.67 + 0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:10 2011