| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,614,099 – 7,614,153 |
| Length | 54 |
| Max. P | 0.827818 |

| Location | 7,614,099 – 7,614,153 |
|---|---|
| Length | 54 |
| Sequences | 8 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 83.65 |
| Shannon entropy | 0.29016 |
| G+C content | 0.32549 |
| Mean single sequence MFE | -10.00 |
| Consensus MFE | -7.76 |
| Energy contribution | -8.01 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
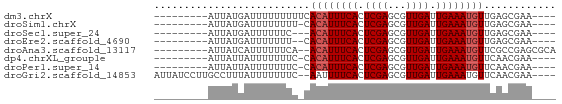
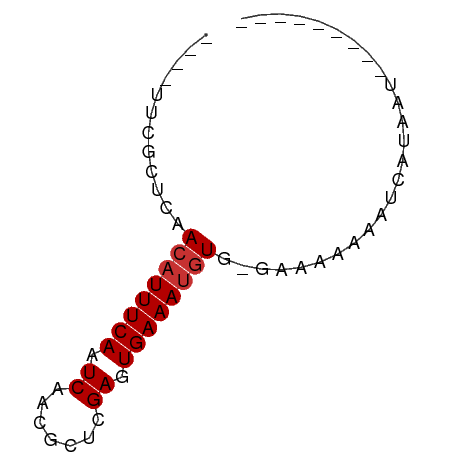
>dm3.chrX 7614099 54 + 22422827 ---------AUUAUGAUUUUUUUUUCACAUUUCACUCGAGCGUUGAUUGAAAUGUUGAGCGAA---- ---------..............(((((((((((.((((...)))).))))))).))))....---- ( -10.40, z-score = -1.10, R) >droSim1.chrX 6087309 53 + 17042790 ---------AUUAUGAUUUUUUUU-CACAUUUCACUCGAGCGUUGAUUGAAAUGUUGAGCGAA---- ---------.............((-(((((((((.((((...)))).))))))).))))....---- ( -10.40, z-score = -1.10, R) >droSec1.super_24 819140 51 + 912625 ---------AUUAUGAUUUUUUC---ACAUUUCACUCGAGCGUUGAUUGAAAUGUUGAGCGAA---- ---------.......(((.(((---((((((((.((((...)))).))))))).)))).)))---- ( -10.50, z-score = -1.10, R) >droEre2.scaffold_4690 16533574 52 + 18748788 ---------AUUAUGAUUUUUUU--CACAUUUCACUCGAGCGUUGAUUGAAAUGUUGAGCGAA---- ---------............((--(((((((((.((((...)))).))))))).))))....---- ( -10.40, z-score = -1.08, R) >droAna3.scaffold_13117 443337 56 + 5790199 ---------AUUAUCAUUUUUUCA--ACAUUUCACUCGAGCGUUGAUUGAAAUGUUCGCCGAGCGCA ---------..............(--((((((((.((((...)))).))))))))).((.....)). ( -10.40, z-score = -0.56, R) >dp4.chrXL_group1e 11632363 53 + 12523060 ---------AUUAUUAUUUUUUUC-CACAUUUCACUCGAGCGUUGAUUGAAAUGUUCAACGAA---- ---------...............-.((((((((.((((...)))).))))))))........---- ( -8.80, z-score = -1.69, R) >droPer1.super_14 1088968 53 + 2168203 ---------AUUAUUAUUUUUUUC-CACAUUUCACUCGAGCGUUGAUUGAAAUGUUCAACGAA---- ---------...............-.((((((((.((((...)))).))))))))........---- ( -8.80, z-score = -1.69, R) >droGri2.scaffold_14853 1800079 61 - 10151454 AUUAUCCUUGCCUUUAUUUUUUUC--AAUUUUCACUCGAGCGUUGAUUGAAAUGUUCAACGAA---- ....((.(((....((((((..((--(((.(((....))).)))))..))))))..))).)).---- ( -10.30, z-score = -2.14, R) >consensus _________AUUAUGAUUUUUUUC_CACAUUUCACUCGAGCGUUGAUUGAAAUGUUCAGCGAA____ ..........................((((((((.((((...)))).))))))))............ ( -7.76 = -8.01 + 0.25)
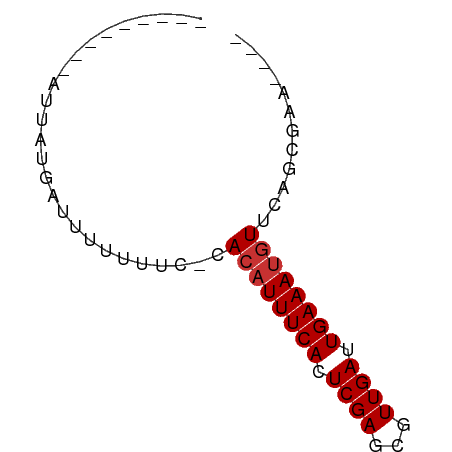
| Location | 7,614,099 – 7,614,153 |
|---|---|
| Length | 54 |
| Sequences | 8 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 83.65 |
| Shannon entropy | 0.29016 |
| G+C content | 0.32549 |
| Mean single sequence MFE | -8.98 |
| Consensus MFE | -7.31 |
| Energy contribution | -7.56 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7614099 54 - 22422827 ----UUCGCUCAACAUUUCAAUCAACGCUCGAGUGAAAUGUGAAAAAAAAAUCAUAAU--------- ----.....(((.(((((((.((.......)).))))))))))...............--------- ( -9.10, z-score = -2.30, R) >droSim1.chrX 6087309 53 - 17042790 ----UUCGCUCAACAUUUCAAUCAACGCUCGAGUGAAAUGUG-AAAAAAAAUCAUAAU--------- ----.....(((.(((((((.((.......)).)))))))))-)..............--------- ( -9.10, z-score = -2.25, R) >droSec1.super_24 819140 51 - 912625 ----UUCGCUCAACAUUUCAAUCAACGCUCGAGUGAAAUGU---GAAAAAAUCAUAAU--------- ----.....(((.(((((((.((.......)).))))))))---))............--------- ( -9.10, z-score = -2.09, R) >droEre2.scaffold_4690 16533574 52 - 18748788 ----UUCGCUCAACAUUUCAAUCAACGCUCGAGUGAAAUGUG--AAAAAAAUCAUAAU--------- ----.....(((.(((((((.((.......)).)))))))))--).............--------- ( -9.10, z-score = -2.17, R) >droAna3.scaffold_13117 443337 56 - 5790199 UGCGCUCGGCGAACAUUUCAAUCAACGCUCGAGUGAAAUGU--UGAAAAAAUGAUAAU--------- ..(((((((((..............))).))))))......--...............--------- ( -11.64, z-score = -0.92, R) >dp4.chrXL_group1e 11632363 53 - 12523060 ----UUCGUUGAACAUUUCAAUCAACGCUCGAGUGAAAUGUG-GAAAAAAAUAAUAAU--------- ----........((((((((.((.......)).)))))))).-...............--------- ( -8.50, z-score = -1.29, R) >droPer1.super_14 1088968 53 - 2168203 ----UUCGUUGAACAUUUCAAUCAACGCUCGAGUGAAAUGUG-GAAAAAAAUAAUAAU--------- ----........((((((((.((.......)).)))))))).-...............--------- ( -8.50, z-score = -1.29, R) >droGri2.scaffold_14853 1800079 61 + 10151454 ----UUCGUUGAACAUUUCAAUCAACGCUCGAGUGAAAAUU--GAAAAAAAUAAAGGCAAGGAUAAU ----.((.(((....(((((((...(((....)))...)))--))))..........))).)).... ( -6.84, z-score = -0.14, R) >consensus ____UUCGCUCAACAUUUCAAUCAACGCUCGAGUGAAAUGUG_GAAAAAAAUCAUAAU_________ ............((((((((.((.......)).)))))))).......................... ( -7.31 = -7.56 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:07 2011