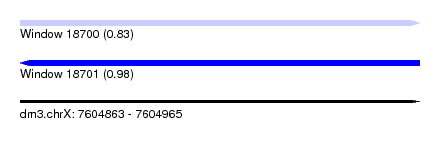
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,604,863 – 7,604,965 |
| Length | 102 |
| Max. P | 0.978641 |

| Location | 7,604,863 – 7,604,965 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 62.17 |
| Shannon entropy | 0.66692 |
| G+C content | 0.39083 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -10.84 |
| Energy contribution | -13.24 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7604863 102 + 22422827 -AUUGAU--AAAGAAAUAUUUUGAACGGCGCGCCAAUUAAGUGCUACCCAUGCGGGC-AAUUUAA-CAUGGCUAGCAUUGACCGUCGAUAAA-UUUUUAAAUGCUCGA -......--..((((((...((((.(((...........(((((((.(((((.....-.......-))))).)))))))..)))))))...)-))))).......... ( -19.22, z-score = 0.75, R) >droSim1.chrX_random 2316601 106 + 5698898 -AUUGAUUAAAAAAAAUAUUUUGAACGGCGCGCCAAUUCAGUGCUACCCAUGAAGAC-AAUAUAAGUAUGGGCAGCAUUGGCCGUCGAUAAUCUUUUUAAAUGCUCGA -.((((((((((......)))))).(((((.((((.....(((((.((((((.....-........)))))).)))))))))))))).................)))) ( -26.92, z-score = -1.78, R) >droSec1.super_24 810021 106 + 912625 -AUUGAUUAAAAAAAAUAUUUUGAACGGGGCGCCAAUUCUGUGCUACCCAUGAAGAC-AAUAUAAGUAUGGGUAGCAUUGGCCGUCGAUAAUCUUUUUAAAUGCUCGA -.((((((((((......)))))).(((.(.((((.....((((((((((((.....-........))))))))))))))))).))).................)))) ( -27.62, z-score = -2.09, R) >droEre2.scaffold_4690 16524244 101 + 18748788 -UUUGAUAAAAAAAAACAUUUUGAAAAUCGCGCCAAUUAAGUGCGACCCAUGCCUCCGAAUGUACGUAUGGGCAGCAUUGGCUAACGAUUAA-CUUUUAAAUG----- -...............(((((.((((((((.((((.....((((..(((((((............)))))))..))))))))...)))))..-.))).)))))----- ( -24.50, z-score = -2.41, R) >droVir3.scaffold_12970 3882613 84 + 11907090 CAAUUAUUUCGGCAAACUUUUCACGACGCGCCUCUUAUUAUUUUGAAAUGCGCGGGC-AAAGCACGUGCUGCAAACUUUGCUUGA----------------------- ..........((((((.(((.((((.(((((...(((......)))...))))).((-...)).))))....))).))))))...----------------------- ( -17.20, z-score = 0.45, R) >consensus _AUUGAUUAAAAAAAAUAUUUUGAACGGCGCGCCAAUUAAGUGCUACCCAUGCAGAC_AAUAUAAGUAUGGGCAGCAUUGGCCGUCGAUAAA_UUUUUAAAUGCUCGA .........................(((((.((((.....(((((.((((((..............)))))).))))))))))))))..................... (-10.84 = -13.24 + 2.40)

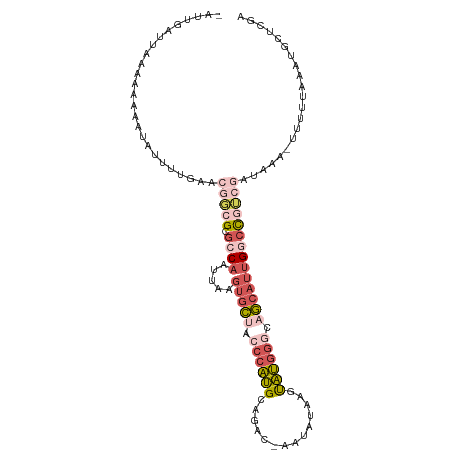
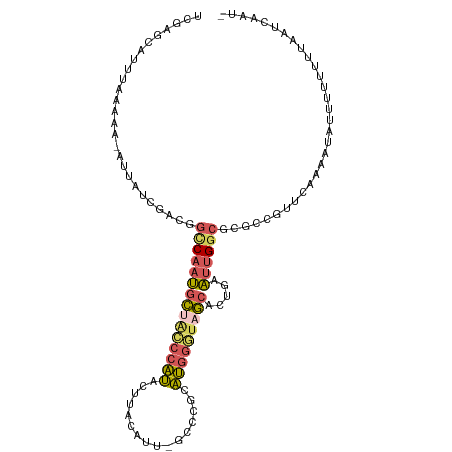
| Location | 7,604,863 – 7,604,965 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 62.17 |
| Shannon entropy | 0.66692 |
| G+C content | 0.39083 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -12.11 |
| Energy contribution | -12.51 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7604863 102 - 22422827 UCGAGCAUUUAAAAA-UUUAUCGACGGUCAAUGCUAGCCAUG-UUAAAUU-GCCCGCAUGGGUAGCACUUAAUUGGCGCGCCGUUCAAAAUAUUUCUUU--AUCAAU- ..((((...(((...-.)))..(.((((((((((((.(((((-(......-....)))))).))))).....))))).)).))))).............--......- ( -22.20, z-score = -0.49, R) >droSim1.chrX_random 2316601 106 - 5698898 UCGAGCAUUUAAAAAGAUUAUCGACGGCCAAUGCUGCCCAUACUUAUAUU-GUCUUCAUGGGUAGCACUGAAUUGGCGCGCCGUUCAAAAUAUUUUUUUUAAUCAAU- ........(((((((((.(((((.(((((((((((((((((((.......-))....)))))))))).....))))).)).))......))).))))))))).....- ( -27.50, z-score = -2.34, R) >droSec1.super_24 810021 106 - 912625 UCGAGCAUUUAAAAAGAUUAUCGACGGCCAAUGCUACCCAUACUUAUAUU-GUCUUCAUGGGUAGCACAGAAUUGGCGCCCCGUUCAAAAUAUUUUUUUUAAUCAAU- ........(((((((((.(((.(((((((((((((((((((((.......-))....)))))))))).....)))))....))))....))).))))))))).....- ( -26.10, z-score = -2.91, R) >droEre2.scaffold_4690 16524244 101 - 18748788 -----CAUUUAAAAG-UUAAUCGUUAGCCAAUGCUGCCCAUACGUACAUUCGGAGGCAUGGGUCGCACUUAAUUGGCGCGAUUUUCAAAAUGUUUUUUUUUAUCAAA- -----.........(-..((((((..((((((((.((((((.(.(........).).)))))).))).....)))))))))))..).....................- ( -23.90, z-score = -1.57, R) >droVir3.scaffold_12970 3882613 84 - 11907090 -----------------------UCAAGCAAAGUUUGCAGCACGUGCUUU-GCCCGCGCAUUUCAAAAUAAUAAGAGGCGCGUCGUGAAAAGUUUGCCGAAAUAAUUG -----------------------....(((((.(((...(((((((((((-((....))(((.......)))..))))))))).))..))).)))))........... ( -19.40, z-score = 0.10, R) >consensus UCGAGCAUUUAAAAA_AUUAUCGACGGCCAAUGCUACCCAUACUUACAUU_GCCCGCAUGGGUAGCACUGAAUUGGCGCGCCGUUCAAAAUAUUUUUUUUAAUCAAU_ ..........................(((((((((((((((................))))))))).....))))))............................... (-12.11 = -12.51 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:05 2011