
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,585,778 – 7,585,860 |
| Length | 82 |
| Max. P | 0.751898 |

| Location | 7,585,778 – 7,585,860 |
|---|---|
| Length | 82 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 61.29 |
| Shannon entropy | 0.74711 |
| G+C content | 0.48705 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -6.63 |
| Energy contribution | -7.57 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
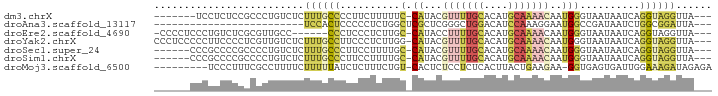
>dm3.chrX 7585778 82 + 22422827 ---UAACCUACCUGAUUAUUACCCAUUGUUUUGCAUGUGCAAAACGUAUG-GAAAAAGAAGGGGCAAAGAGACAGGGCGGAGAGGA------- ---...((..((((........((((((((((((....)))))))).)))-).....(......).......))))..))......------- ( -20.20, z-score = -1.59, R) >droAna3.scaffold_13117 418701 65 + 5790199 ---UAAUCCGCCAGAUUAUCGGCCAUUCCUUUGGAUGUCCAGCCCGAGCGAGCCAGAGGGGGAGUGGA------------------------- ---(((((.....)))))....(((((((.((((....))))(((...(......).)))))))))).------------------------- ( -19.30, z-score = 0.25, R) >droEre2.scaffold_4690 16505032 82 + 18748788 ---UAACCUACCUGAUUAUUACCCAUUGUUUUGCAUGUGCAAAAGGUAUG-GCAAGAGGGAGGG------GGCAACGCGAGACAGGGAGGGG- ---...(((.((((.......(((.((.((((((.(((((.....)))))-)))))).)).)))------(....)......)))).)))..- ( -26.80, z-score = -2.57, R) >droYak2.chrX 8035681 89 - 21770863 ---UAACCUACCUGAUUAUUACCCAUUGUUUUGCAUGUGCAAAACGUAUG-CCAAGAGGGAAGGCAAAGAGACAACGAGGGGAAGGGGGAGGG ---...(((.(((..((....(((...(((((((....)))))))...((-((.........))))............))).))..)))))). ( -26.10, z-score = -2.25, R) >droSec1.super_24 791593 83 + 912625 ---UAACCUACCUGAUUAUUACCCAUUGUUUUGCAUGUGCAAAACGUAUG-GCAAAAGGAAGGGCAAAGAGACAGGGGCGGGGCGGG------ ---...(((.((((.((.((.(((.((.((((((.(((((.....)))))-)))))).)).))).)).))..))))...))).....------ ( -24.20, z-score = -1.37, R) >droSim1.chrX 6066434 83 + 17042790 ---UAACCUACCUGAUUAUUACCCAUUGUUUUGCAUGUGCAAAACGUAUG-GCAAAAGGAAGGGCAAAGAGACAGGGGCGGGGCGGG------ ---...(((.((((.((.((.(((.((.((((((.(((((.....)))))-)))))).)).))).)).))..))))...))).....------ ( -24.20, z-score = -1.37, R) >droMoj3.scaffold_6500 22346920 82 - 32352404 UCUCUAUCUUUCCAAUCACUCACC-UUCUUCAGUAAGUGAGAGGAGAGUG-ACAGAAAGAGAUAAAAAGAAAAGGCGAAAGGGA--------- (((((.((((((...((((((.((-((..(((.....))))))).)))))-)..))))))........(......)...)))))--------- ( -20.40, z-score = -0.83, R) >consensus ___UAACCUACCUGAUUAUUACCCAUUGUUUUGCAUGUGCAAAACGUAUG_GCAAAAGGAAGGGCAAAGAGACAGCGAGGGGGAGGG______ .....................(((.((.((((((.(((((.....))))).)))))).)).)))............................. ( -6.63 = -7.57 + 0.94)



| Location | 7,585,778 – 7,585,860 |
|---|---|
| Length | 82 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 61.29 |
| Shannon entropy | 0.74711 |
| G+C content | 0.48705 |
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -4.88 |
| Energy contribution | -6.59 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7585778 82 - 22422827 -------UCCUCUCCGCCCUGUCUCUUUGCCCCUUCUUUUUC-CAUACGUUUUGCACAUGCAAAACAAUGGGUAAUAAUCAGGUAGGUUA--- -------......((..((((.....((((((.((.......-.....(((((((....))))))))).))))))....))))..))...--- ( -20.90, z-score = -3.53, R) >droAna3.scaffold_13117 418701 65 - 5790199 -------------------------UCCACUCCCCCUCUGGCUCGCUCGGGCUGGACAUCCAAAGGAAUGGCCGAUAAUCUGGCGGAUUA--- -------------------------(((........((..((((....))))..))(((((...)).)))((((......)))))))...--- ( -19.00, z-score = 0.16, R) >droEre2.scaffold_4690 16505032 82 - 18748788 -CCCCUCCCUGUCUCGCGUUGCC------CCCUCCCUCUUGC-CAUACCUUUUGCACAUGCAAAACAAUGGGUAAUAAUCAGGUAGGUUA--- -..(((.((((......((((((------(.....(....).-......((((((....))))))....)))))))...)))).)))...--- ( -19.00, z-score = -1.70, R) >droYak2.chrX 8035681 89 + 21770863 CCCUCCCCCUUCCCCUCGUUGUCUCUUUGCCUUCCCUCUUGG-CAUACGUUUUGCACAUGCAAAACAAUGGGUAAUAAUCAGGUAGGUUA--- .......(((...(((.(((((..(((((((.........))-))...(((((((....)))))))...)))..))))).))).)))...--- ( -22.40, z-score = -2.67, R) >droSec1.super_24 791593 83 - 912625 ------CCCGCCCCGCCCCUGUCUCUUUGCCCUUCCUUUUGC-CAUACGUUUUGCACAUGCAAAACAAUGGGUAAUAAUCAGGUAGGUUA--- ------......((...((((.....((((((........(.-....)(((((((....)))))))...))))))....))))..))...--- ( -21.70, z-score = -2.10, R) >droSim1.chrX 6066434 83 - 17042790 ------CCCGCCCCGCCCCUGUCUCUUUGCCCUUCCUUUUGC-CAUACGUUUUGCACAUGCAAAACAAUGGGUAAUAAUCAGGUAGGUUA--- ------......((...((((.....((((((........(.-....)(((((((....)))))))...))))))....))))..))...--- ( -21.70, z-score = -2.10, R) >droMoj3.scaffold_6500 22346920 82 + 32352404 ---------UCCCUUUCGCCUUUUCUUUUUAUCUCUUUCUGU-CACUCUCCUCUCACUUACUGAAGAA-GGUGAGUGAUUGGAAAGAUAGAGA ---------................(((((((((.((((.((-(((((.(((((((.....)).)).)-)).))))))).))))))))))))) ( -24.30, z-score = -2.36, R) >consensus ______CCCUCCCCCCCCCUGUCUCUUUGCCCUUCCUUUUGC_CAUACGUUUUGCACAUGCAAAACAAUGGGUAAUAAUCAGGUAGGUUA___ ..................(((.....((((((........(......)(((((((....)))))))...))))))....)))........... ( -4.88 = -6.59 + 1.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:24:03 2011