
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,548,274 – 10,548,379 |
| Length | 105 |
| Max. P | 0.706320 |

| Location | 10,548,274 – 10,548,379 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.16 |
| Shannon entropy | 0.51785 |
| G+C content | 0.62136 |
| Mean single sequence MFE | -41.46 |
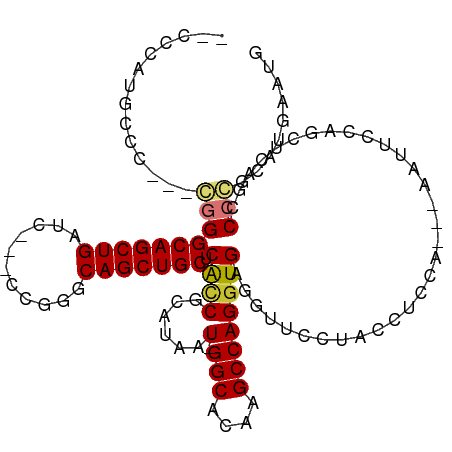
| Consensus MFE | -20.35 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.706320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
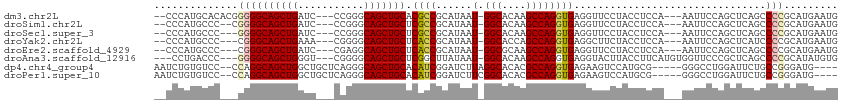
>dm3.chr2L 10548274 105 + 23011544 --CCCAUGCACACGGGGGCAGCUGAUC---CCGGGCAGCUGCACGCCGCAUAAU-GGCACAAGCCAGGUGAGGUUCCUACCUCCA---AAUUCCAGCUCAGCCCCGCAUGAAUG --..(((((....(((((((((((...---.....))))))).....((....(-(((....)))).(.(((((....)))))).---.......))....))))))))).... ( -39.80, z-score = -1.16, R) >droSim1.chr2L 10352441 103 + 22036055 --CCCAUGCCC--CGGGGCAGCUGAUC---CCGGGCAGCUGCUCGCCGCAUAAU-GGCACAAGCCAGGUGAGGUUCCUACCUCCA---AAUUCCAGCUCAGCCCCGCAUGAAUG --..(((((..--.((((((((((...---((((((....)))).........(-(((....)))))).(((((....)))))..---.....)))))..)))))))))).... ( -41.40, z-score = -2.04, R) >droSec1.super_3 5974779 102 + 7220098 --CCCAUGCCC---GGGGCAGCUGAUC---CCGGGCAGCUGCUCGCCGCAUAAU-GGCACAAGCCAGGUGAGGUUCCUACCUCCA---AAUUCCAGCUCAGCCCCGCAUGAAUG --..(((((..---((((((((((...---((((((....)))).........(-(((....)))))).(((((....)))))..---.....)))))..)))))))))).... ( -41.80, z-score = -2.14, R) >droYak2.chr2L 6957420 102 + 22324452 --CCCAUGCCC---CGGGCAGCUGAAA---CGGGGCAGCUGCUCACCGCAUAAU-GGCACCAGCCAGGUGAGGCUUCUACCUCCA---AAUUCCAGCUCAUCCCCGCAUGAAUG --..(((((..---.(((.(((((...---.((((.((..(((((((......(-(((....))))))))).))..)).))))..---.....)))))...))).))))).... ( -35.30, z-score = -0.94, R) >droEre2.scaffold_4929 11762089 102 - 26641161 --CCCAUGCCC---CGGGCAGCUGAUC---CGAGGCAGCUGCUCACCGCAUAAU-GGCGCAAGCCAGGUGAGGUUCCUACCUCCA---AAUUCCAGCUCAGCCCCGCAUGAAUG --..(((((..---.(((((((((...---.((((.((..(((((((......(-(((....))))))))).))..)).))))..---.....)))))..)))).))))).... ( -41.80, z-score = -3.12, R) >droAna3.scaffold_12916 14398834 104 + 16180835 ---CCUGACCC---GGGGCAGCUGGGU---CGGGGCAGCUGCUCGGCUUAUAAU-GGCACAAGCCAGGUGAGGUACUUACCUUCAUGUGGUUCCCGCUCAGCCCCGCAUAUGUG ---((((((((---((.....))))))---))))(((..(((..((((......-(((...(((((.(((((((....))).)))).)))))...))).))))..)))..))). ( -44.60, z-score = -1.17, R) >dp4.chr4_group4 2754979 103 + 6586962 AAUCUGUGUCC--CCAGGCAGCUGGCUGCUCAGGGCAGCUGCACAUCGGAUCUUAGGCACACGCCAGGUGAGAAGUCCAUGCG-----GGGCCUGGAUUCUGCCGGGAUG---- .......((((--(..((((((.((((((.....)))))))).....((((((.((((...(((..((........))..)))-----..))))))))))))))))))).---- ( -43.70, z-score = -0.60, R) >droPer1.super_10 1763273 103 + 3432795 AAUCUGUGUCC--CCAGGCAGCUGGCUGCUCAGGGCAGCUGCACAUCGGAUCUUCGGCACACGCCAGGUGAGAAGUCCAUGCG-----GGGCCUGGAUUCUGCCGGGAUG---- .........((--((..(((((((.((.....)).))))))).(((.(((.((((..(((.(....)))).)))))))))).)-----)))(((((......)))))...---- ( -43.30, z-score = -0.36, R) >consensus __CCCAUGCCC___CGGGCAGCUGAUC___CCGGGCAGCUGCUCACCGCAUAAU_GGCACAAGCCAGGUGAGGUUCCUACCUCCA___AAUUCCAGCUCAGCCCCGCAUGAAUG ..............((((((((((.((.....)).))))))).............(((....)))(((((.(....))))))....................)))......... (-20.35 = -20.85 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:27 2011