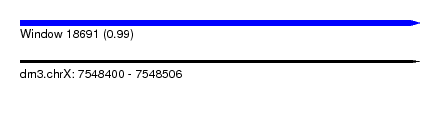
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,548,400 – 7,548,506 |
| Length | 106 |
| Max. P | 0.992210 |

| Location | 7,548,400 – 7,548,506 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.72 |
| Shannon entropy | 0.65435 |
| G+C content | 0.46750 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -10.53 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7548400 106 + 22422827 UUUAACCUCUUGACAAGCCCC-CCGACUUCC---UUUGACCCCUUAAGUAGCUUUGUGUGUUGCAAUUGCAGACACCUGCUUA---ACCUCCUAUUUUUCAAAAAUUUCACUC--- .........((((........-.(((.....---.))).....((((((((....((((.((((....)))))))))))))))---)...........))))...........--- ( -13.90, z-score = -1.33, R) >droPer1.super_14 1008810 100 + 2168203 -GUAACGCUCUG------CCCCCAGACCCUG---ACCCACUGCUUAAGUAGCUUUAUGUGUUGCACUUGCAGACACCUGCUUA---ACCCACCAAUGAACCCGCACCACAUUU--- -(((((((((((------....)))).....---.....((((....))))......)))))))....((((....))))...---...........................--- ( -17.90, z-score = -1.80, R) >dp4.chrXL_group1e 11554336 100 + 12523060 -GUAACGCUCUG------CCCCCAGACCCUG---ACCCACUGCUUAAGUAGCUUUAUGUGUUGCACUUGCAGACACCUGCUUA---ACCCACCAAUGAACCCGCACCACAUUU--- -(((((((((((------....)))).....---.....((((....))))......)))))))....((((....))))...---...........................--- ( -17.90, z-score = -1.80, R) >droAna3.scaffold_13117 387518 96 + 5790199 -------------------UGGCAAGUAGCUGG-GGCACCCCCCUUAGUAGCUUUGUGUAUUGCAGUUGCAGACACCUGCCCCGAUAGCCGGGAAUAUUCGGAAAACACAAAGCCC -------------------.(((.....)))((-((....))))......(((((((((.((.(....((((....))))((((.....)))).......).)).))))))))).. ( -33.10, z-score = -1.60, R) >droEre2.scaffold_4690 16465256 106 + 18748788 -CUAACCUCUGGAAAAAAACAACGGGCUUCC---UUUGACCCCUUAAGUAGCUUUGUGUGUUGCAGCUGCAAACACCUGCUUA---ACCUCCUCUUUUUCAAAAAUUUCACUC--- -........(((((((.......((((....---...).))).((((((((....((((.((((....)))))))))))))))---).......)))))))............--- ( -20.70, z-score = -1.52, R) >droYak2.chrX 7998158 105 - 21770863 -UUAACCUCUUGUUGAGCCAA-CGUGCUUCC---UUUGACCCCUUAAGUAGCUUUGUGUGUUGCAGUUGCAGACACCUGCUUA---ACCUCCAAUUUUUCACGAUUUUCACUU--- -.....(((.....)))....-((((.....---.(((.....((((((((....((((.((((....)))))))))))))))---)....))).....))))..........--- ( -19.70, z-score = -1.42, R) >droSec1.super_24 755663 109 + 912625 UUUAACCUCUUGACAAGCCCC-CGGACUUCCUCCUUUGACCCCUUAAGUAGCUUUGUGUGUUGCAAUUGCAGACACCUGCUUA---ACCUCCUACUUUUCAAAAAUUUCACUC--- .........((((.(((....-.(((.....))).........((((((((....((((.((((....)))))))))))))))---).......))).))))...........--- ( -18.80, z-score = -2.23, R) >droSim1.chrX 6036839 109 + 17042790 UUUAACCUCUUGACAAGCCCC-CGGACUUCCUCCUUUGACCCCUUAAGUAGCUUUGUGUGUUGCAAUUGCAGACACCUGCUUA---ACCUCCUACUUUUCAAAAAUUUCACUC--- .........((((.(((....-.(((.....))).........((((((((....((((.((((....)))))))))))))))---).......))).))))...........--- ( -18.80, z-score = -2.23, R) >consensus _UUAACCUCUUGA_AAGCCCC_CGGACUUCC___UUUGACCCCUUAAGUAGCUUUGUGUGUUGCAAUUGCAGACACCUGCUUA___ACCUCCUAAUUUUCAAAAAUUUCACUC___ ............................................(((((((....((((.((((....)))))))))))))))................................. (-10.53 = -11.05 + 0.52)

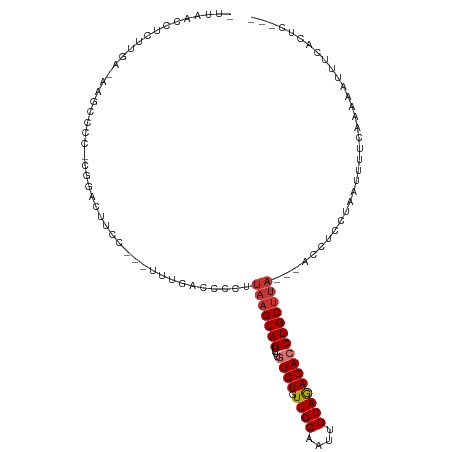
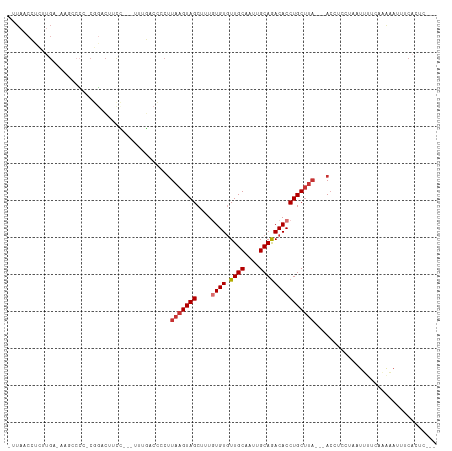
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:57 2011