
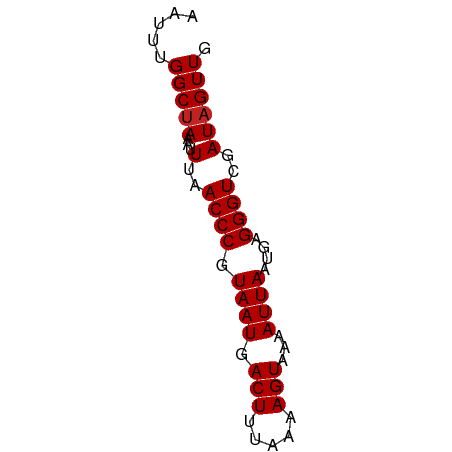
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,539,514 – 7,539,575 |
| Length | 61 |
| Max. P | 0.830180 |

| Location | 7,539,514 – 7,539,575 |
|---|---|
| Length | 61 |
| Sequences | 11 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 99.70 |
| Shannon entropy | 0.00720 |
| G+C content | 0.29657 |
| Mean single sequence MFE | -10.39 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.19 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743049 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
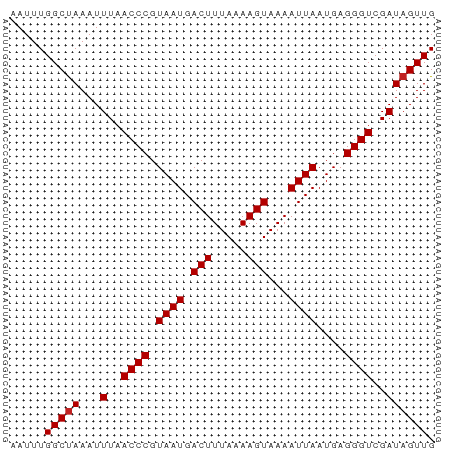
>dm3.chrX 7539514 61 + 22422827 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droSim1.chrX_random 2303929 61 + 5698898 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droSec1.super_24 746791 61 + 912625 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droYak2.chrX 7988119 61 - 21770863 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droEre2.scaffold_4690 16456137 61 + 18748788 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droAna3.scaffold_13117 380789 61 + 5790199 AAUUUGGCCAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG ........(((..(..((((.((((.(((.....)))...))))....))))..)...))) ( -9.30, z-score = -0.65, R) >dp4.chrXL_group1e 11543979 61 + 12523060 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droWil1.scaffold_180777 1538560 61 - 4753960 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droVir3.scaffold_12970 3811449 61 + 11907090 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droMoj3.scaffold_6473 8307686 61 + 16943266 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >droGri2.scaffold_15203 1900608 61 - 11997470 AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). ( -10.50, z-score = -1.47, R) >consensus AAUUUGGCUAAAUUUAACCCGUAAUGACUUUAAAAGUAAAAUUAAUGAGGGUCGAUAGUUG .....(((((...(..((((.((((.(((.....)))...))))....))))..)))))). (-10.10 = -10.19 + 0.09)

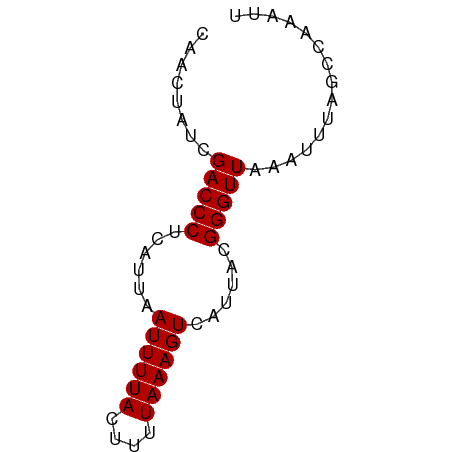
| Location | 7,539,514 – 7,539,575 |
|---|---|
| Length | 61 |
| Sequences | 11 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 99.70 |
| Shannon entropy | 0.00720 |
| G+C content | 0.29657 |
| Mean single sequence MFE | -6.90 |
| Consensus MFE | -6.90 |
| Energy contribution | -6.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830180 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 7539514 61 - 22422827 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droSim1.chrX_random 2303929 61 - 5698898 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droSec1.super_24 746791 61 - 912625 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droYak2.chrX 7988119 61 + 21770863 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droEre2.scaffold_4690 16456137 61 - 18748788 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droAna3.scaffold_13117 380789 61 - 5790199 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUGGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -0.53, R) >dp4.chrXL_group1e 11543979 61 - 12523060 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droWil1.scaffold_180777 1538560 61 + 4753960 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droVir3.scaffold_12970 3811449 61 - 11907090 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droMoj3.scaffold_6473 8307686 61 - 16943266 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >droGri2.scaffold_15203 1900608 61 + 11997470 CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90, z-score = -1.39, R) >consensus CAACUAUCGACCCUCAUUAAUUUUACUUUUAAAGUCAUUACGGGUUAAAUUUAGCCAAAUU ........(((((......((((((....))))))......)))))............... ( -6.90 = -6.90 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:55 2011