| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,480,839 – 7,480,931 |
| Length | 92 |
| Max. P | 0.961815 |

| Location | 7,480,839 – 7,480,931 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 71.91 |
| Shannon entropy | 0.59891 |
| G+C content | 0.35362 |
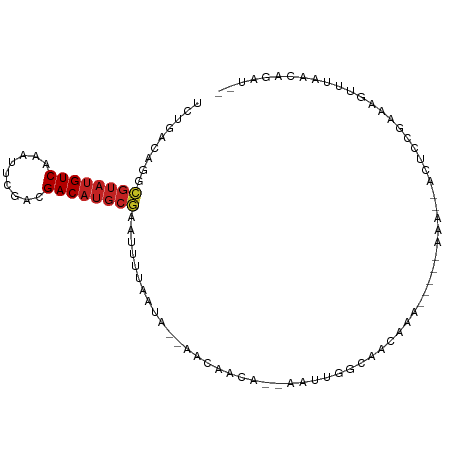
| Mean single sequence MFE | -15.27 |
| Consensus MFE | -8.07 |
| Energy contribution | -8.37 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
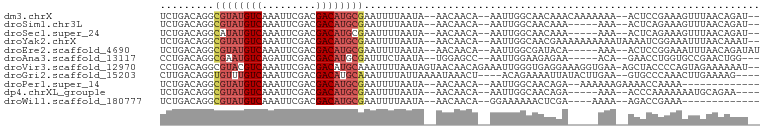
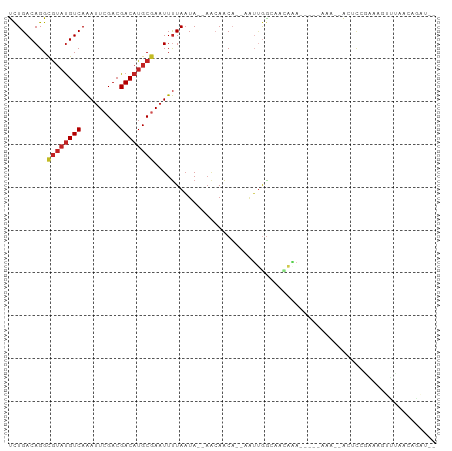
>dm3.chrX 7480839 92 + 22422827 UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--AAUUGGCAACAAACAAAAAAA--ACUCCGAAAGUUUAACAGAU-- ((((.....((((((((.........))))))))..........--.......--..(((....))).......((--(((.....)))))..)))).-- ( -17.50, z-score = -2.53, R) >droSim1.chr3L 13201997 87 - 22553184 UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--AAUUGGCAACAAA-----AAA--ACUCAGAAAGUUUAACAGAU-- (((((....((((((((.........))))))))..........--.......--..(((....))).-----...--..))))).............-- ( -17.80, z-score = -2.92, R) >droSec1.super_24 688652 87 + 912625 UCUGACAGGCAUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--AAUUGGCAACAAA-----AAA--ACUCAGAAAGUUUAACAGAU-- (((((...((((.((((......))))..))))...........--.......--..(((....))).-----...--..))))).............-- ( -14.40, z-score = -2.14, R) >droYak2.chrX 7923038 94 - 21770863 UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--AAUUGGCAACGAAAAAAAAAAUAAAAUCGGAAAUUUAACAAAU-- (((((....((((((((.........))))))))..........--.......--..(((....)))..............)))))............-- ( -17.00, z-score = -2.53, R) >droEre2.scaffold_4690 16395622 89 + 18748788 UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--AAUUGGCGAUACA-----AAA--ACUCCGGAAAUUUAACAGAUAU ((((.....((((((((.........))))))))..........--......(--((((..((.....-----...--....))..)))))..))))... ( -14.60, z-score = -1.21, R) >droAna3.scaffold_13117 328787 86 + 5790199 CCUGACAGGCGAAUGUCAGAUUCGACGACAUGCGAUUUCUAAUA--UGGAGCC--AAUUGGAAGAGAA-----ACA--GAACCUGGUGCCGAACUGG--- .((((((......)))))).((((.(.((......((((((((.--((....)--)))))))))....-----.((--(...)))))).))))....--- ( -17.60, z-score = 0.04, R) >droVir3.scaffold_12970 3720905 97 + 11907090 CCUGACAGGCGUACGUCAAAUUCGACGACAUGCAAAUUUUAAUAGUAACAACAGAAAUUGGGUGAGGAAAGGUGAA-AGCUACCCCAGUAGAAAAAAU-- (((.((..((((.((((......))))..))))...............(((......))).)).)))...((((..-...))))..............-- ( -16.10, z-score = -0.01, R) >droGri2.scaffold_15203 1819328 90 - 11997470 CUUGACAGGUGUUUGUCAAAUUCGACGACAUGCAAAUUUUAUUAAAAUAAACU----ACAGAAAAUUAUACUUGAA--GUGCCCAAACUUGAAAAG---- .....((((((((((((......))))).........(((((....)))))..----..........)))))))((--((......))))......---- ( -10.70, z-score = 0.21, R) >droPer1.super_14 925424 81 + 2168203 UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--AAUUGGCAACAGA--AAAAAAGAAAACCAAAA------------- .......((((((((((.........))))))))..........--.......--..(((....))).--...........))....------------- ( -14.20, z-score = -2.29, R) >dp4.chrXL_group1e 11476207 85 + 12523060 UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--AAUUGGCAACAGA-----AAA--ACCCAAAAAAAUGCAGAA---- ((((.((..((((((((.........))))))))..........--.......--..(((....))).-----...--...........)))))).---- ( -17.30, z-score = -2.66, R) >droWil1.scaffold_180777 3303063 77 + 4753960 UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA--AACAACA--GGAAAAAACUCGA----AAAA--AGACCGAAA------------- .......((((((((((.........))))))))..........--.......--.((......))..----....--...))....------------- ( -10.80, z-score = -0.45, R) >consensus UCUGACAGGCGUAUGUCAAAUUCGACGACAUGCGAAUUUUAAUA__AACAACA__AAUUGGCAACAAA_____AAA__ACUCCGAAAGUUUAACAGAU__ .........((((((((.........)))))))).................................................................. ( -8.07 = -8.37 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:49 2011