
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,475,887 – 7,475,981 |
| Length | 94 |
| Max. P | 0.960874 |

| Location | 7,475,887 – 7,475,979 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Shannon entropy | 0.15626 |
| G+C content | 0.32527 |
| Mean single sequence MFE | -19.79 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
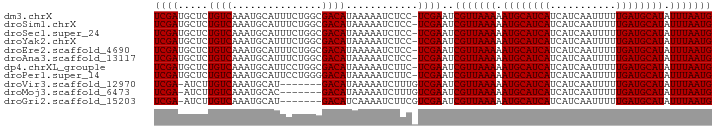
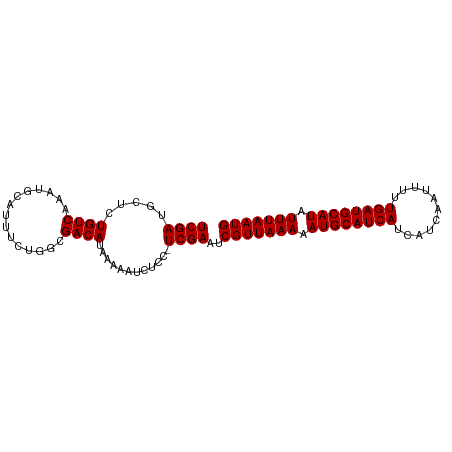
>dm3.chrX 7475887 92 + 22422827 UCGAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........))).....((.((........)).))-))))..(((((((.((((((((...........)))))))).))))))) ( -19.90, z-score = -2.53, R) >droSim1.chrX 6003830 92 + 17042790 UCGAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........))).....((.((........)).))-))))..(((((((.((((((((...........)))))))).))))))) ( -19.90, z-score = -2.53, R) >droSec1.super_24 684012 92 + 912625 UCGAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........))).....((.((........)).))-))))..(((((((.((((((((...........)))))))).))))))) ( -19.90, z-score = -2.53, R) >droYak2.chrX 7917601 92 - 21770863 UCGAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........))).....((.((........)).))-))))..(((((((.((((((((...........)))))))).))))))) ( -19.90, z-score = -2.53, R) >droEre2.scaffold_4690 16390524 92 + 18748788 UCGAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........))).....((.((........)).))-))))..(((((((.((((((((...........)))))))).))))))) ( -19.90, z-score = -2.53, R) >droAna3.scaffold_13117 323303 92 + 5790199 UCGAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........))).....((.((........)).))-))))..(((((((.((((((((...........)))))))).))))))) ( -19.90, z-score = -2.53, R) >dp4.chrXL_group1e 11470920 92 + 12523060 UCGAUGCUCUGUCAAAUGCAUUCCUGGCGACAUAAAAAUCUUC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........)))....((....))...........-))))..(((((((.((((((((...........)))))))).))))))) ( -19.10, z-score = -2.23, R) >droPer1.super_14 920145 92 + 2168203 UCGAUGCUCUGUCAAAUGCAUUCCUGGGGACAUAAAAAUCUUC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG (((((((..........))).....(((((........)))))-))))..(((((((.((((((((...........)))))))).))))))) ( -20.70, z-score = -2.62, R) >droVir3.scaffold_12970 3711646 85 + 11907090 UCGA-AUCUUGUCAAAUGCAU-------GACAUAAAAAUCUUUGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG ((((-....(((((......)-------))))............))))..(((((((.((((((((...........)))))))).))))))) ( -19.59, z-score = -2.81, R) >droMoj3.scaffold_6473 8214366 85 + 16943266 UCGA-AUCUUGUCAAAUGCAC-------GACAUAAAAAUCUUUGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG ....-...............(-------((((..........)))))...(((((((.((((((((...........)))))))).))))))) ( -19.30, z-score = -2.79, R) >droGri2.scaffold_15203 1814155 85 - 11997470 UCGA-AUCUUGUCAAAUGCAU-------GACAUCAAAAUCUUCGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG ((((-....(((((......)-------))))............))))..(((((((.((((((((...........)))))))).))))))) ( -19.59, z-score = -2.93, R) >consensus UCGAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC_UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUG ((((.....((((...............))))............))))..(((((((.((((((((...........)))))))).))))))) (-16.01 = -16.01 + -0.00)
| Location | 7,475,889 – 7,475,981 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Shannon entropy | 0.17444 |
| G+C content | 0.31416 |
| Mean single sequence MFE | -19.04 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.91 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
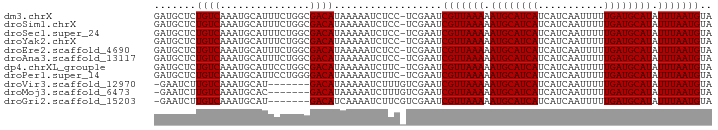
>dm3.chrX 7475889 92 + 22422827 GAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))...((.((........)).))-......(((((((.((((((((...........)))))))).))))))).. ( -19.00, z-score = -2.09, R) >droSim1.chrX 6003832 92 + 17042790 GAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))...((.((........)).))-......(((((((.((((((((...........)))))))).))))))).. ( -19.00, z-score = -2.09, R) >droSec1.super_24 684014 92 + 912625 GAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))...((.((........)).))-......(((((((.((((((((...........)))))))).))))))).. ( -19.00, z-score = -2.09, R) >droYak2.chrX 7917603 92 - 21770863 GAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))...((.((........)).))-......(((((((.((((((((...........)))))))).))))))).. ( -19.00, z-score = -2.09, R) >droEre2.scaffold_4690 16390526 92 + 18748788 GAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))...((.((........)).))-......(((((((.((((((((...........)))))))).))))))).. ( -19.00, z-score = -2.09, R) >droAna3.scaffold_13117 323305 92 + 5790199 GAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))...((.((........)).))-......(((((((.((((((((...........)))))))).))))))).. ( -19.00, z-score = -2.09, R) >dp4.chrXL_group1e 11470922 92 + 12523060 GAUGCUCUGUCAAAUGCAUUCCUGGCGACAUAAAAAUCUUC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))..((....))...........-......(((((((.((((((((...........)))))))).))))))).. ( -18.20, z-score = -1.78, R) >droPer1.super_14 920147 92 + 2168203 GAUGCUCUGUCAAAUGCAUUCCUGGGGACAUAAAAAUCUUC-UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA (((((..........)))))..((((((.((....)).)))-)))...(((((((.((((((((...........)))))))).))))))).. ( -20.10, z-score = -2.25, R) >droVir3.scaffold_12970 3711648 85 + 11907090 -GAAUCUUGUCAAAUGCAU-------GACAUAAAAAUCUUUGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA -......(((((......)-------))))..................(((((((.((((((((...........)))))))).))))))).. ( -18.60, z-score = -2.33, R) >droMoj3.scaffold_6473 8214368 85 + 16943266 -GAAUCUUGUCAAAUGCAC-------GACAUAAAAAUCUUUGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA -.................(-------((((..........)))))...(((((((.((((((((...........)))))))).))))))).. ( -19.90, z-score = -2.98, R) >droGri2.scaffold_15203 1814157 85 - 11997470 -GAAUCUUGUCAAAUGCAU-------GACAUCAAAAUCUUCGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA -......(((((......)-------))))..................(((((((.((((((((...........)))))))).))))))).. ( -18.60, z-score = -2.38, R) >consensus GAUGCUCUGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUCC_UCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUA .......((((...............))))..................(((((((.((((((((...........)))))))).))))))).. (-15.91 = -15.91 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:48 2011