
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,465,839 – 7,465,935 |
| Length | 96 |
| Max. P | 0.995597 |

| Location | 7,465,839 – 7,465,935 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.42326 |
| G+C content | 0.40711 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
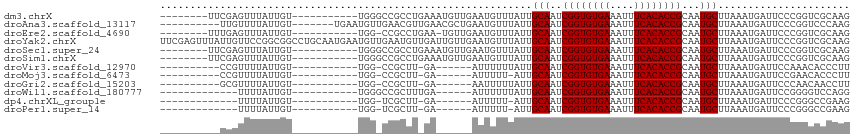
>dm3.chrX 7465839 96 + 22422827 --------UUCGAGUUUAUUGU-----------UGGGCCGCCUGAAAUGUUGAAUGUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGUCGCAAG --------.............(-----------(((((((.(......)..(((((((((..(((..((((((((....))))))))...))).))))).)))).))))).))). ( -25.10, z-score = -1.10, R) >droAna3.scaffold_13117 312616 98 + 5790199 ----------UUGUUUUAUUGU-------UGAAUGUUGAACGUUGAACGCUGAAUGUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGUCCCAAG ----------..........((-------(.((((.....)))).)))((((((((((((..(((..((((((((....))))))))...))).))))).))))..)))...... ( -24.40, z-score = -1.85, R) >droEre2.scaffold_4690 16380859 94 + 18748788 --------UUUGAGUUUAUUGU-----------UGG-CCGCCUGAA-UGUUGAAUGUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGUCGCAAG --------..........((((-----------.((-(((...(((-(......((((((..(((..((((((((....))))))))...))).)))))))))).))))))))). ( -25.80, z-score = -1.86, R) >droYak2.chrX 7905604 115 - 21770863 UUCGAGUUUAUUGUUCCGGCGGCCUGCAAUGAAUGUUGAAUGUUGAUUGUUGAAUGUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGUCGCAAG ..................(((((((((((((((((((.((((.....)))).)))))))))))))..((((((((....))))))))...................))))))... ( -38.10, z-score = -3.33, R) >droSec1.super_24 673796 96 + 912625 --------UUCGAGUUUAUUGU-----------UGGGCCGCCUGAAAUGUUGAAUGUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGUCGCAAG --------.............(-----------(((((((.(......)..(((((((((..(((..((((((((....))))))))...))).))))).)))).))))).))). ( -25.10, z-score = -1.10, R) >droSim1.chrX 5993478 96 + 17042790 --------UUCGAGUUUAUUGU-----------UGGGCCGCCUGAAAUGUUGAAUGUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGUCGCAAG --------.............(-----------(((((((.(......)..(((((((((..(((..((((((((....))))))))...))).))))).)))).))))).))). ( -25.10, z-score = -1.10, R) >droVir3.scaffold_12970 3697038 86 + 11907090 ----------CCGUUUUAUUGU-----------UGG-CCGCUU-GA------AUUUUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCAAACACCCUU ----------............-----------.((-......-((------(((.((((..(((..((((((((....))))))))...))).)))).)))))......))... ( -20.40, z-score = -2.38, R) >droMoj3.scaffold_6473 8202578 85 + 16943266 ----------CCGUUUUAUUGU-----------UGG-CCGCUU-GA------AUUUUU-AUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCGAACACCCUU ----------............-----------((.-.((...-((------(((...-...(((..((((((((....))))))))...)))......)))))))..))..... ( -19.90, z-score = -1.99, R) >droGri2.scaffold_15203 1803557 86 - 11997470 ----------GCGUUUUAUUGU-----------UGG-CCGCUU-GA------AAUUUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCAACAACCUU ----------........((((-----------(((-......-((------(...((((..(((..((((((((....))))))))...))).))))...)))))))))).... ( -23.00, z-score = -2.88, R) >droWil1.scaffold_180777 3320337 85 + 4753960 -------------UUUUAUUGU-----------UGGGCCGCUUUGA------AUUUUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCGGGGUCCAGG -------------........(-----------((((((.(...((------(((.((((..(((..((((((((....))))))))...))).)))).))))).).))))))). ( -31.80, z-score = -4.31, R) >dp4.chrXL_group1e 11457955 82 + 12523060 -------------UUUUAUUGU-----------UGG-UCGCUU-GA------AUUUUU-AUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGGCCGAAG -------------........(-----------(((-((.(..-((------(((...-...(((..((((((((....))))))))...)))......)))))..))))))).. ( -26.20, z-score = -3.34, R) >droPer1.super_14 907022 82 + 2168203 -------------UUUUAUUGU-----------UGG-UCGCUU-GA------AUUUUU-AUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGGCCGAAG -------------........(-----------(((-((.(..-((------(((...-...(((..((((((((....))))))))...)))......)))))..))))))).. ( -26.20, z-score = -3.34, R) >consensus __________CGGUUUUAUUGU___________UGG_CCGCUU_GA______AAUGUUUAUUGCAAUCGGUGUGAAAUUUCACACCGCAAUGCUUAAAUGAUUCCCGGUCCCAAG ..............................................................(((..((((((((....))))))))...)))...................... (-15.10 = -15.10 + -0.00)
| Location | 7,465,839 – 7,465,935 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.42326 |
| G+C content | 0.40711 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
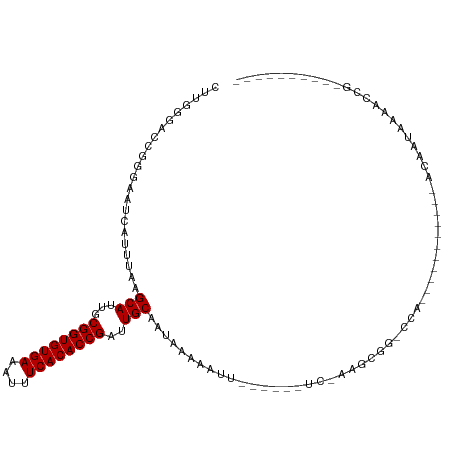
>dm3.chrX 7465839 96 - 22422827 CUUGCGACCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAACAUUCAACAUUUCAGGCGGCCCA-----------ACAAUAAACUCGAA-------- ........((((..........(((...((((((((....))))))))..)))....................((....)).-----------........))))..-------- ( -22.30, z-score = -1.41, R) >droAna3.scaffold_13117 312616 98 - 5790199 CUUGGGACCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAACAUUCAGCGUUCAACGUUCAACAUUCA-------ACAAUAAAACAA---------- ....(((((..((((.......(((...((((((((....))))))))..))).......)))).).))))...............-------............---------- ( -22.34, z-score = -2.02, R) >droEre2.scaffold_4690 16380859 94 - 18748788 CUUGCGACCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAACAUUCAACA-UUCAGGCGG-CCA-----------ACAAUAAACUCAAA-------- .(((.(.(((.((((.......(((...((((((((....))))))))..))).......))))..(.-....).)))-)))-----------).............-------- ( -20.74, z-score = -1.35, R) >droYak2.chrX 7905604 115 + 21770863 CUUGCGACCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAACAUUCAACAAUCAACAUUCAACAUUCAUUGCAGGCCGCCGGAACAAUAAACUCGAA ((.(((.((..((((.......(((...((((((((....))))))))..))).......))))................((.....))..)).))).))............... ( -24.84, z-score = -1.07, R) >droSec1.super_24 673796 96 - 912625 CUUGCGACCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAACAUUCAACAUUUCAGGCGGCCCA-----------ACAAUAAACUCGAA-------- ........((((..........(((...((((((((....))))))))..)))....................((....)).-----------........))))..-------- ( -22.30, z-score = -1.41, R) >droSim1.chrX 5993478 96 - 17042790 CUUGCGACCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAACAUUCAACAUUUCAGGCGGCCCA-----------ACAAUAAACUCGAA-------- ........((((..........(((...((((((((....))))))))..)))....................((....)).-----------........))))..-------- ( -22.30, z-score = -1.41, R) >droVir3.scaffold_12970 3697038 86 - 11907090 AAGGGUGUUUGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAAAAAU------UC-AAGCGG-CCA-----------ACAAUAAAACGG---------- ..((.((((((((.........(((...((((((((....))))))))..))).........)------))-))))).-)).-----------............---------- ( -28.37, z-score = -4.48, R) >droMoj3.scaffold_6473 8202578 85 - 16943266 AAGGGUGUUCGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAU-AAAAAU------UC-AAGCGG-CCA-----------ACAAUAAAACGG---------- ..((.((((..((((..((...(((...((((((((....))))))))..)))...-))..))------))-.)))).-)).-----------............---------- ( -25.00, z-score = -3.33, R) >droGri2.scaffold_15203 1803557 86 + 11997470 AAGGUUGUUGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAAAAUU------UC-AAGCGG-CCA-----------ACAAUAAAACGC---------- ..(((((((.(((((..((...(((...((((((((....))))))))..)))...))..)))------))-.)))))-)).-----------............---------- ( -28.70, z-score = -4.29, R) >droWil1.scaffold_180777 3320337 85 - 4753960 CCUGGACCCCGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAAAAAU------UCAAAGCGGCCCA-----------ACAAUAAAA------------- ..(((.((.(.((((..((((.(((...((((((((....))))))))..)))..))))..))------))...).)).)))-----------.........------------- ( -25.40, z-score = -3.60, R) >dp4.chrXL_group1e 11457955 82 - 12523060 CUUCGGCCCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAU-AAAAAU------UC-AAGCGA-CCA-----------ACAAUAAAA------------- ....(((.(..((((..((...(((...((((((((....))))))))..)))...-))..))------))-..).).-)).-----------.........------------- ( -21.80, z-score = -2.66, R) >droPer1.super_14 907022 82 - 2168203 CUUCGGCCCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAU-AAAAAU------UC-AAGCGA-CCA-----------ACAAUAAAA------------- ....(((.(..((((..((...(((...((((((((....))))))))..)))...-))..))------))-..).).-)).-----------.........------------- ( -21.80, z-score = -2.66, R) >consensus CUUGGGACCGGGAAUCAUUUAAGCAUUGCGGUGUGAAAUUUCACACCGAUUGCAAUAAAAAUU______UC_AAGCGG_CCA___________ACAAUAAAACCG__________ ......................(((...((((((((....))))))))..))).............................................................. (-17.60 = -17.60 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:46 2011