| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,464,586 – 7,464,639 |
| Length | 53 |
| Max. P | 0.939471 |

| Location | 7,464,586 – 7,464,639 |
|---|---|
| Length | 53 |
| Sequences | 7 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Shannon entropy | 0.46783 |
| G+C content | 0.60515 |
| Mean single sequence MFE | -21.91 |
| Consensus MFE | -10.89 |
| Energy contribution | -11.73 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
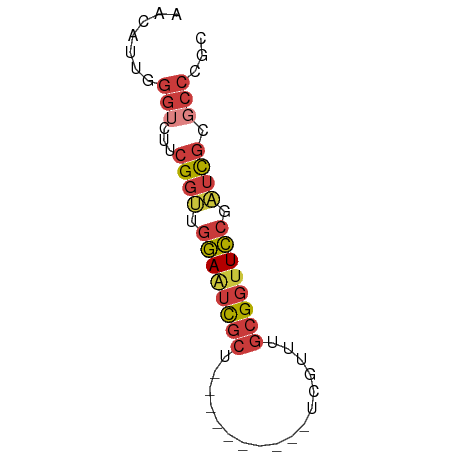
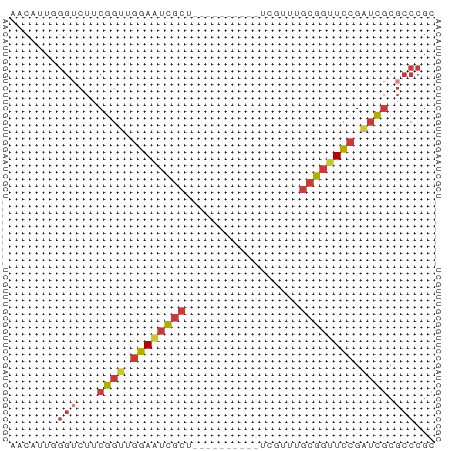
>dm3.chrX 7464586 53 + 22422827 AACAUUGGGUCUUCGGUUGGAAUCGCU----------UCGUUUGCGGUUCCGAUCGCGCCCGC ......((((...(((((((((((((.----------......))))))))))))).)))).. ( -26.80, z-score = -4.36, R) >droSim1.chrX 5992237 53 + 17042790 AACAUUGGGUCUUCGGUUGGAAUCGCU----------UCGUUUGCGGUUCCGAUCGCGCCCGC ......((((...(((((((((((((.----------......))))))))))))).)))).. ( -26.80, z-score = -4.36, R) >droSec1.super_24 672552 53 + 912625 AACAUUGGGUCUUCGGUUGGAAUCGGU----------UCGUUUGCGGUUCCGAUUGCGCCCGC ......((((...((((((((((((..----------.......)))))))))))).)))).. ( -20.30, z-score = -1.99, R) >droEre2.scaffold_4690 16379389 53 + 18748788 AACAUUGGGUCUUCGGUUGGAAUCGCU----------UCGUUUGCGGUUCCGAUCGCGCCCGC ......((((...(((((((((((((.----------......))))))))))))).)))).. ( -26.80, z-score = -4.36, R) >droAna3.scaffold_13117 311574 53 + 5790199 AACAUUCGGUCUUCGGCGGGAAUCGCU----------CCGGUUGCGGUUCCGAUCGCGCCCGC ..............((((((((((((.----------......)))))))).....))))... ( -17.60, z-score = 0.46, R) >dp4.chrXL_group1e 11456597 63 + 12523060 AACAUUCGGACUUCGGUGCGAGUCGCUGCUUGCUGCUUCGGUUGCGGUUCCGAUCGCGCCCGC ..............(((((((.(((..(((.((.((....)).)))))..))))))))))... ( -23.80, z-score = -0.77, R) >droVir3.scaffold_12970 3695446 50 + 11907090 ACGAGUGCGACUUCGCCAGCAACUCCGACU-------CUGACUGCGACUGCGGUUCC------ ..(((((((....))).....)))).....-------..((((((....))))))..------ ( -11.30, z-score = 0.41, R) >consensus AACAUUGGGUCUUCGGUUGGAAUCGCU__________UCGUUUGCGGUUCCGAUCGCGCCCGC .......(((...((((.((((((((.................)))))))).)))).)))... (-10.89 = -11.73 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:44 2011