| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,452,038 – 7,452,179 |
| Length | 141 |
| Max. P | 0.946099 |

| Location | 7,452,038 – 7,452,179 |
|---|---|
| Length | 141 |
| Sequences | 7 |
| Columns | 164 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Shannon entropy | 0.38707 |
| G+C content | 0.51031 |
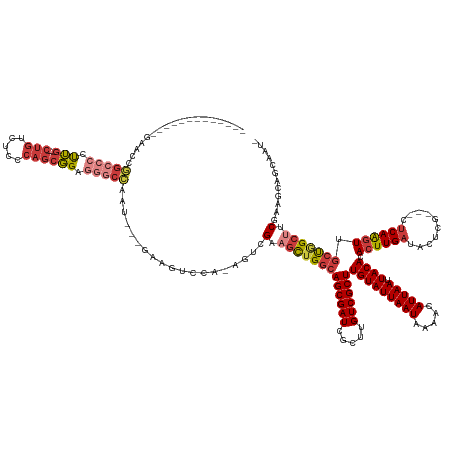
| Mean single sequence MFE | -46.89 |
| Consensus MFE | -24.40 |
| Energy contribution | -26.14 |
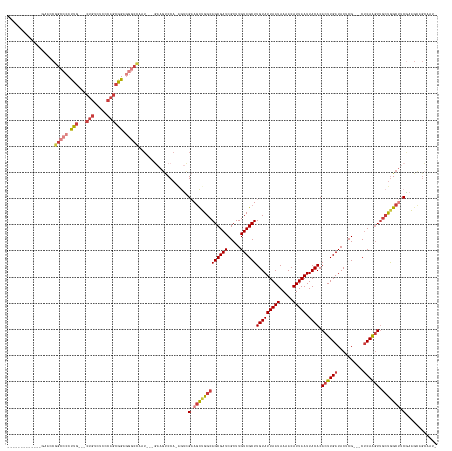
| Covariance contribution | 1.74 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
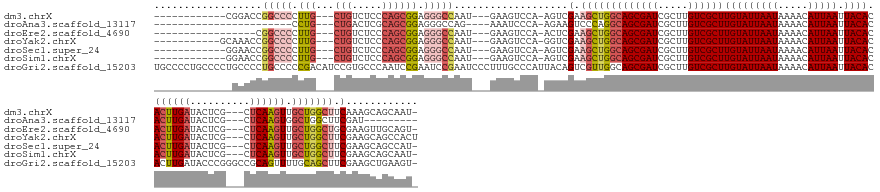
>dm3.chrX 7452038 141 - 22422827 ------------CGGACCGGCCCCUUG---CUGUCUCCCAGCGGAGGGCCAAU---GAAGUCCA-AGUCGAAGCUGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACUCG---CUCAAGUUGCUGGCUUCAAAGCAGCAAU- ------------.((((.(((((..((---(((.....)))))..)))))...---...)))).-....(((((..((((((((.....))))).(((((((((.....))))).)))).((((((......---.)))))))))..)))))...........- ( -49.80, z-score = -2.91, R) >droAna3.scaffold_13117 300416 121 - 5790199 -----------------------CCUG---CUGACUCGCAGCGGAGGGCCAG----AAAUCCCA-AGAAGUCCCAGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACUCG---CUCAAGUGGCUGGCUUCGAU--------- -----------------------.(((---(((.....)))))).(((....----....))).-.((((((...(((((((((.....))))))(((((((((.....))))).))))(((((((......---.))))))))))))))))...--------- ( -37.40, z-score = -1.67, R) >droEre2.scaffold_4690 16365898 136 - 18748788 -----------------CGGCCCCUUG---CUGUCUCCCAGCGGAGGGCCAAU---GAAGUCCA-ACUCGAAGCUGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACUCG---CUCAAGUUGCUGGCUGCGAAGUUGCAGU- -----------------.(((((..((---(((.....)))))..)))))...---......((-(((((.(((..((((((((.....))))).(((((((((.....))))).)))).((((((......---.)))))))))..))).)).)))))....- ( -49.20, z-score = -2.85, R) >droYak2.chrX 7889807 143 + 21770863 -----------GCAAACCGGCCCCUUG---CUGUCUCCCAGCGGAGGGCCAAU---GAAGUCCA-GGUCGAAGCUGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACUCG---CUCAAGUUGCUGGCUUCGAAGCAGCCACU -----------((...(((((((..((---(((.....)))))..)))))..(---(.....))-))(((((((..((((((((.....))))).(((((((((.....))))).)))).((((((......---.)))))))))..))))))).))....... ( -52.60, z-score = -3.22, R) >droSec1.super_24 660005 141 - 912625 ------------GGAACCGGCCCCUUG---CUGUCUCCCAGCGGAGGGCCAAU---GAAGUCCA-AGUCGAAGCUGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACUCG---CUCAAGUUGCUGGCUUCGAAGCAGCCAU- ------------((....(((((..((---(((.....)))))..)))))...---........-..(((((((..((((((((.....))))).(((((((((.....))))).)))).((((((......---.)))))))))..))))))).....))..- ( -51.60, z-score = -3.23, R) >droSim1.chrX 5978470 141 - 17042790 ------------GGAACCGGCCCCUUG---CUGUCUCCCAGCGGAGGGCCAAU---GAAGUCCA-AGUCGAAGCUGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACUCG---CUCAAGUUGCUGGCUUCGAAGCAGCAAU- ------------(((...(((((..((---(((.....)))))..)))))...---....))).-..(((((((..((((((((.....))))).(((((((((.....))))).)))).((((((......---.)))))))))..))))))).........- ( -51.20, z-score = -3.19, R) >droGri2.scaffold_15203 1789008 163 + 11997470 UGCCCCUGCCCCUGCCCCUGCCCCCGACAUCCGUGCCCAAUCCGAAUCCGAAUCCCUUUGCCCAUUACAGUCGUUGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACCCGGGCCGCAGUUUUGCAGCUUCGAAGCUGAAGU- (((..((((....((((.((((..((((....(((..(((...((.......))...)))..)))....))))..))))(((((.....))))).(((((((((.....))))).))))............)))).))))....)))((((((....))))))- ( -36.40, z-score = -0.58, R) >consensus _____________GAACCGGCCCCUUG___CUGUCUCCCAGCGGAGGGCCAAU___GAAGUCCA_AGUCGAAGCUGGCAGCGAUCGCUUGUCGCUUGUAUUAAUAAAACAUUAAUUACACACUUGAUACUCG___CUCAAGUUGCUGGCUUCGAAGCAGCAAU_ ..................(((((.(((...(((.....)))))).)))))...................(.(((((((((((((.....))))))(((((((((.....))))).)))).((((((..........)))))).))))))).)............ (-24.40 = -26.14 + 1.74)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:43 2011