
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,445,204 – 7,445,301 |
| Length | 97 |
| Max. P | 0.788790 |

| Location | 7,445,204 – 7,445,301 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.41109 |
| G+C content | 0.35194 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.45 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
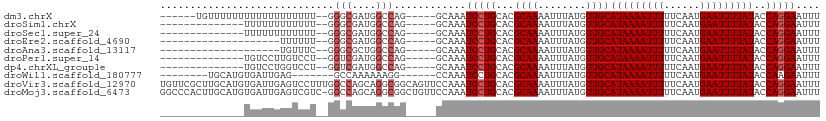
>dm3.chrX 7445204 97 + 22422827 ------UGUUUUUUUUUUUUUUUUUU--GGGCGAUGGCCAG-----GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU ------....................--.(((....)))..-----.....(((((...((((........))))(((((((((......)))))))))..))))).... ( -18.40, z-score = -0.28, R) >droSim1.chrX 5971724 89 + 17042790 --------------UUUUUUUUUUUU--GGGCGAUGGCCAG-----GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU --------------............--.(((....)))..-----.....(((((...((((........))))(((((((((......)))))))))..))))).... ( -18.40, z-score = -0.87, R) >droSec1.super_24 653229 89 + 912625 --------------UUUUUUUUUUUU--GGGCGAUGGCCAG-----GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU --------------............--.(((....)))..-----.....(((((...((((........))))(((((((((......)))))))))..))))).... ( -18.40, z-score = -0.87, R) >droEre2.scaffold_4690 16359029 83 + 18748788 --------------------UUUUUU--GGGCGAUGGCCAG-----GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU --------------------......--.(((....)))..-----.....(((((...((((........))))(((((((((......)))))))))..))))).... ( -18.40, z-score = -1.02, R) >droAna3.scaffold_13117 294993 83 + 5790199 --------------------UGUUUC--GGGCGCUGGCCAG-----GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU --------------------((((..--.(((....))).)-----)))..(((((...((((........))))(((((((((......)))))))))..))))).... ( -19.80, z-score = -1.02, R) >droPer1.super_14 884146 89 + 2168203 --------------UGUCCUUGUCCU--GGUCGAUGGCCAG-----GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU --------------.....(((.(((--(((.....)))))-----)))).(((((...((((........))))(((((((((......)))))))))..))))).... ( -26.40, z-score = -3.31, R) >dp4.chrXL_group1e 11435354 89 + 12523060 --------------UGUCCUGGUCCU--GGUCGAUGGCCAG-----GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU --------------..((((((((((--(((.....)))))-----)............((((........))))(((((((((......)))))))))))))))).... ( -28.00, z-score = -3.33, R) >droWil1.scaffold_180777 1415092 89 - 4753960 --------UGCAUGUGAUUGAG-------GCCAAAAAAGG------CCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAAGAAUUU --------((((.(.((((..(-------(((......))------)).))))))))).((((........))))(((((((((......)))))))))........... ( -21.50, z-score = -2.30, R) >droVir3.scaffold_12970 3672797 110 + 11907090 UGUUCGCUUGCAUGUGAUUGAGUCCUUUGGCCAGCAGGCGGCAGUUCCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU (((.(((((((..((.(..(....)..).))..))))))))))........(((((...((((........))))(((((((((......)))))))))..))))).... ( -27.40, z-score = -0.77, R) >droMoj3.scaffold_6473 8176811 109 + 16943266 GGCCCACUUGCAUGUGAUUGAGUCGUC-GGCCAGCAGGCGGCUGUUCCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU ((..(((......)))....(((((((-........)))))))...))...(((((...((((........))))(((((((((......)))))))))..))))).... ( -26.90, z-score = -0.43, R) >consensus ______________UGUUUUUGUUUU__GGGCGAUGGCCAG_____GCAAAUCCUGCACGCAAAAUUUAUGUUGCAUAAAAUUUUUCAAUGAAUUUUAUACCAGGAAUUU .............................(((....)))............(((((...((((........))))(((((((((......)))))))))..))))).... (-15.05 = -15.45 + 0.40)


| Location | 7,445,204 – 7,445,301 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.41109 |
| G+C content | 0.35194 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.550782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7445204 97 - 22422827 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC-----CUGGCCAUCGCCC--AAAAAAAAAAAAAAAAAACA------ ....(((((((((((((((........))))))))))..(((.........))))))))((((.-----..(((....))))--))).................------ ( -15.60, z-score = -0.25, R) >droSim1.chrX 5971724 89 - 17042790 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC-----CUGGCCAUCGCCC--AAAAAAAAAAAA-------------- ....(((((((((((((((........))))))))))..(((.........))))))))((((.-----..(((....))))--))).........-------------- ( -15.60, z-score = -0.40, R) >droSec1.super_24 653229 89 - 912625 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC-----CUGGCCAUCGCCC--AAAAAAAAAAAA-------------- ....(((((((((((((((........))))))))))..(((.........))))))))((((.-----..(((....))))--))).........-------------- ( -15.60, z-score = -0.40, R) >droEre2.scaffold_4690 16359029 83 - 18748788 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC-----CUGGCCAUCGCCC--AAAAAA-------------------- ....(((((((((((((((........))))))))))..(((.........))))))))((((.-----..(((....))))--)))...-------------------- ( -15.60, z-score = -0.42, R) >droAna3.scaffold_13117 294993 83 - 5790199 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC-----CUGGCCAGCGCCC--GAAACA-------------------- ...(((...((((((((((........))))))))))............((((.((((.....)-----)))....))))..--)))...-------------------- ( -16.70, z-score = -0.24, R) >droPer1.super_14 884146 89 - 2168203 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC-----CUGGCCAUCGACC--AGGACAAGGACA-------------- ....((((.((((((((((........))))))))))....(((((((((....)))))))))(-----((((.......))--)))...))))..-------------- ( -23.20, z-score = -2.10, R) >dp4.chrXL_group1e 11435354 89 - 12523060 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC-----CUGGCCAUCGACC--AGGACCAGGACA-------------- ....(((((((...(((((((((....)))..(((((((........)))))))..)))))).(-----((((.......))--))))))))))..-------------- ( -26.60, z-score = -3.03, R) >droWil1.scaffold_180777 1415092 89 + 4753960 AAAUUCUUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGG------CCUUUUUUGGC-------CUCAAUCACAUGCA-------- .........((((((((((........))))))))))...........((((((...((((.((------((......)))-------)..)))).))))))-------- ( -23.90, z-score = -2.83, R) >droVir3.scaffold_12970 3672797 110 - 11907090 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGGAACUGCCGCCUGCUGGCCAAAGGACUCAAUCACAUGCAAGCGAACA .........((((((((((........)))))))))).........(((((.((((.((((.((..((...(((....)))...))..)).))))...)))).))))).. ( -28.30, z-score = -1.61, R) >droMoj3.scaffold_6473 8176811 109 - 16943266 AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGGAACAGCCGCCUGCUGGCC-GACGACUCAAUCACAUGCAAGUGGGCC ...((((..((((((((((........)))))))))).....((((((((....)))))))))))).....(((..((.((.-...((.....))....)).))..))). ( -26.20, z-score = -0.37, R) >consensus AAAUUCCUGGUAUAAAAUUCAUUGAAAAAUUUUAUGCAACAUAAAUUUUGCGUGCAGGAUUUGC_____CUGGCCAUCGCCC__AAAACAAAAACA______________ .........((((((((((........))))))))))....(((((((((....)))))))))........(((....)))............................. (-13.58 = -13.47 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:42 2011