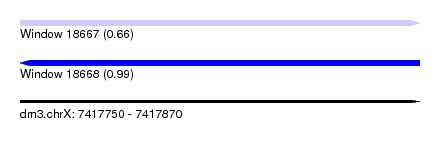
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,417,750 – 7,417,870 |
| Length | 120 |
| Max. P | 0.989994 |

| Location | 7,417,750 – 7,417,870 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.09 |
| Shannon entropy | 0.12590 |
| G+C content | 0.62352 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -30.55 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
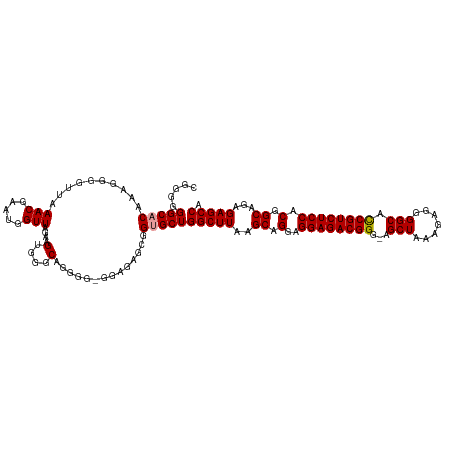
>dm3.chrX 7417750 120 + 22422827 CAGGGGGCACAAAGGGGUUAAACGGACGGUUAGAAGGGGGCAGGGGCGGGGAGGUGCGCUGGCUUAAGCAGGAGGAGACGGGAAGCUAAAGAGGGGCAUCGUCUCCACGGCAGAGAGCCA ...((.((((......(((.....)))............((....))......)))).))(((((..((.(..((((((((...(((.......))).)))))))).).))...))))). ( -35.50, z-score = -1.68, R) >droSec1.super_24 625788 117 + 912625 CGGGGGGCACAAAGGGGUUAAACGAAUGGUUAGAAGUGGGCAGGGA-G-AGAGCGGUGCUGGCUUAAGCAGGAGGAGACGGG-AGCUGAAGAGGGGCACCGUCUCCACGGCAGAGAGCCA ((...(((........)))...))..(((((..((((.((((..(.-.-....)..)))).))))..((.(..((((((((.-.(((.......))).)))))))).).))....))))) ( -36.30, z-score = -1.59, R) >droSim1.chrX 5943076 119 + 17042790 CGGGGGGCACAAAGGGGUUAAACGUAUGGUUAGAAGUGCGCAGGGG-GGAGAGCGGUGCUGGCUUAAGCAGGAGGAGACGGGGAGCUAAAGAGGGGCACCGUCUCCACGGCAGAGAGCCA .....(((...............((((........))))((.(...-(((((.(((((((..(((.(((....(....).....))).)))...)))))))))))).).)).....))). ( -39.30, z-score = -2.15, R) >consensus CGGGGGGCACAAAGGGGUUAAACGAAUGGUUAGAAGUGGGCAGGGG_GGAGAGCGGUGCUGGCUUAAGCAGGAGGAGACGGG_AGCUAAAGAGGGGCACCGUCUCCACGGCAGAGAGCCA .....(((((..........(((.....)))....(....)..............)))))(((((..((.(..((((((((...(((.......))).)))))))).).))...))))). (-30.55 = -30.67 + 0.11)

| Location | 7,417,750 – 7,417,870 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.09 |
| Shannon entropy | 0.12590 |
| G+C content | 0.62352 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.989994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
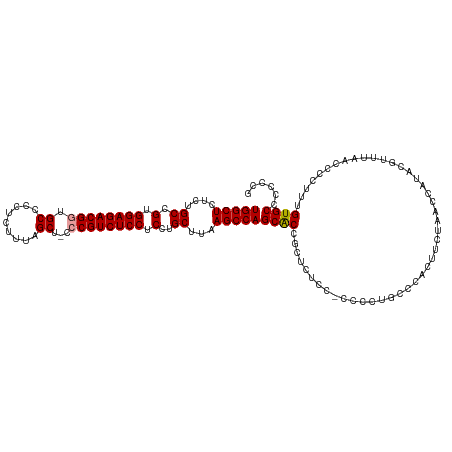
>dm3.chrX 7417750 120 - 22422827 UGGCUCUCUGCCGUGGAGACGAUGCCCCUCUUUAGCUUCCCGUCUCCUCCUGCUUAAGCCAGCGCACCUCCCCGCCCCUGCCCCCUUCUAACCGUCCGUUUAACCCCUUUGUGCCCCCUG (((((....((.(.(((((((..((.........))....))))))).)..))...)))))(((((.......((....))........(((.....))).........)))))...... ( -24.40, z-score = -2.40, R) >droSec1.super_24 625788 117 - 912625 UGGCUCUCUGCCGUGGAGACGGUGCCCCUCUUCAGCU-CCCGUCUCCUCCUGCUUAAGCCAGCACCGCUCU-C-UCCCUGCCCACUUCUAACCAUUCGUUUAACCCCUUUGUGCCCCCCG (((((....((.(.((((((((.((.........)).-.)))))))).)..))...)))))((((.((...-.-.....))........(((.....)))..........))))...... ( -27.60, z-score = -2.90, R) >droSim1.chrX 5943076 119 - 17042790 UGGCUCUCUGCCGUGGAGACGGUGCCCCUCUUUAGCUCCCCGUCUCCUCCUGCUUAAGCCAGCACCGCUCUCC-CCCCUGCGCACUUCUAACCAUACGUUUAACCCCUUUGUGCCCCCCG .(((.....)))..(((((((((((....(((.(((...............))).)))...)))))).)))))-.......((((.........................))))...... ( -29.97, z-score = -3.43, R) >consensus UGGCUCUCUGCCGUGGAGACGGUGCCCCUCUUUAGCU_CCCGUCUCCUCCUGCUUAAGCCAGCACCGCUCUCC_CCCCUGCCCACUUCUAACCAUACGUUUAACCCCUUUGUGCCCCCCG (((((....((.(.((((((((.((.........))...)))))))).)..))...)))))((((.............................................))))...... (-24.17 = -24.28 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:38 2011