
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,409,594 – 7,409,684 |
| Length | 90 |
| Max. P | 0.992536 |

| Location | 7,409,594 – 7,409,684 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Shannon entropy | 0.48867 |
| G+C content | 0.45069 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
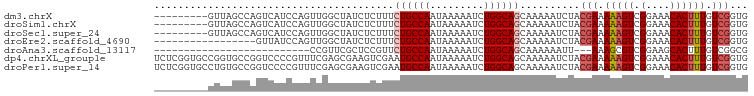
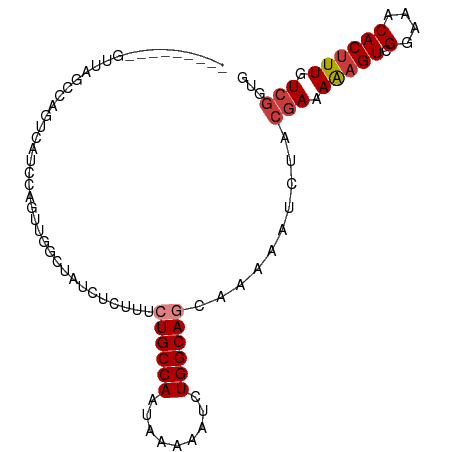
>dm3.chrX 7409594 90 + 22422827 ---------GUUAGCCAGUCAUCCAGUUGGCUAUCUCUUUCUGCCAAUAAAAAUCUGGCAGCAAAAAUCUACGAAAAAGUCGGAAACACUUUGUCGGUG ---------(((.(((((.......((((((...........))))))......)))))))).........(((.(((((.(....)))))).)))... ( -26.32, z-score = -4.23, R) >droSim1.chrX 5935522 90 + 17042790 ---------GUUAGCCAGUCAUCCAGUUGGCUAUCUCUUUCUGCCAAUAAAAAUCUGGCAGCAAAAAUCUACGAAAAAGUCGGAAACACUUUGUCGGUG ---------(((.(((((.......((((((...........))))))......)))))))).........(((.(((((.(....)))))).)))... ( -26.32, z-score = -4.23, R) >droSec1.super_24 617707 90 + 912625 ---------GUUAGCCAGUCAUCCAGUUGGCUAUCUCUUUCUGCCAAUAAAAAUCUGGCAGCAAAAAUCUACGAAAAAGUCGGAAACACUUUGUCGGUG ---------(((.(((((.......((((((...........))))))......)))))))).........(((.(((((.(....)))))).)))... ( -26.32, z-score = -4.23, R) >droEre2.scaffold_4690 16321508 82 + 18748788 -----------------GUUAUCCAGUUGGCUAUCUCUUUCUGCCAAUAAAAAUCUGGCAGCAAAAAUCUACGAAAAAGUCGGAAACACUUUGUCGGUG -----------------(((..(((((((((...........))))).......)))).))).........(((.(((((.(....)))))).)))... ( -18.41, z-score = -2.43, R) >droAna3.scaffold_13117 260725 70 + 5790199 --------------------------CCGUUCGCUCCGUUCUGCCAAUAAAAAUCUGGCAGCAAAAAAUU---AAAGCGUCGGAAGCACUUUGUCGGCG --------------------------.....((((.....((((((.........))))))........(---((((.(.(....)).)))))..)))) ( -15.70, z-score = -0.61, R) >dp4.chrXL_group1e 11395015 99 + 12523060 UCUCGGUGCCGGUGCCGGUCCCCGUUUCGAGCGAAGUCGAAUGCCAAUAAAAAUCUGGCAGCAAAAAUCUACGAAAAAGUCGGAAACACUUUGUCGGUG ....((..(((....)))..))...(((((......)))))(((((.........)))))...........(((.(((((.(....)))))).)))... ( -27.20, z-score = -0.83, R) >droPer1.super_14 843510 99 + 2168203 UCUCGGUGCCUGUGCCGGUCCCCGUUUCGAGCGAAGUCGAAUGCCAAUAAAAAUCUGGCAGCAAAAAUCUACGAAAAAGUCGGAAACACUUUGUCGGUG ....((..((......))..))...(((((......)))))(((((.........)))))...........(((.(((((.(....)))))).)))... ( -23.50, z-score = -0.16, R) >consensus _________GUUAGCCAGUCAUCCAGUUGGCUAUCUCUUUCUGCCAAUAAAAAUCUGGCAGCAAAAAUCUACGAAAAAGUCGGAAACACUUUGUCGGUG ........................................((((((.........))))))..........(((.(((((.(....)))))).)))... (-13.38 = -13.97 + 0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:36 2011