| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,377,494 – 7,377,597 |
| Length | 103 |
| Max. P | 0.992670 |

| Location | 7,377,494 – 7,377,597 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Shannon entropy | 0.42476 |
| G+C content | 0.36970 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -12.14 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.992670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7377494 103 + 22422827 AACCCACCAUCUACUCUC---UCAGUGGGCCAGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUCGGGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC ..(((((...........---...)))))((.(((((....)))))..((((((..------...)))))).....))((((((((((((.....)))))))..)))))... ( -22.34, z-score = -3.14, R) >droEre2.scaffold_4690 16287959 106 + 18748788 AAUCCACCACCACCACCCAACUCAGUGGGCCAGUGAAUGUAUUCAUCAAUGCAAUA------CAUUUGCAUAUUUCGGGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC .........((.((((........))))....(..((((((((((....)).))))------))))..).......))((((((((((((.....)))))))..)))))... ( -26.20, z-score = -3.83, R) >droYak2.chrX 7805051 100 - 21770863 AAUCCACCACCU---AUC---UCAGUGGGCCAGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUAGGGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC ...((.((((..---...---...))))((.((((..((((((....))))))...------)))).)).......))((((((((((((.....)))))))..)))))... ( -22.10, z-score = -2.80, R) >droSec1.super_24 585490 100 + 912625 AACCCACCACCU---CUC---UCAGUGGGCCAGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUCGGGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC ..(((((.....---...---...)))))((.(((((....)))))..((((((..------...)))))).....))((((((((((((.....)))))))..)))))... ( -22.60, z-score = -3.02, R) >droSim1.chrX 5902981 100 + 17042790 AACCCACCACCU---CUC---UCAGUGGGCCAGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUCGGGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC ..(((((.....---...---...)))))((.(((((....)))))..((((((..------...)))))).....))((((((((((((.....)))))))..)))))... ( -22.60, z-score = -3.02, R) >dp4.chrXL_group1e 1159328 95 - 12523060 --------AGGGCAGACC---CCAGUGGGUCUGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUCGAGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC --------...(((((((---(....))))))))..............((((((..------...)))))).......((((((((((((.....)))))))..)))))... ( -26.70, z-score = -3.50, R) >droPer1.super_14 1357499 95 - 2168203 --------AGGGCAGACC---CCAGUGGGUCUGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUCAAGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC --------...(((((((---(....))))))))..............((((((..------...)))))).......((((((((((((.....)))))))..)))))... ( -26.70, z-score = -3.82, R) >droWil1.scaffold_180777 1350598 106 - 4753960 ---UGACUGCUGCUGACU---CAAAUGGGCCAGUGAAUGUAUUCAUCAAUGCAACAAAAACACAUUUGCAUAUUUUGAGCGUAAAAAUUAAUCACUAAUUUUAAUACGAAGC ---(((.(((.((((.((---(....))).))))....))).)))...((((((...........))))))........(((((((((((.....)))))))..)))).... ( -17.20, z-score = 0.30, R) >droVir3.scaffold_12970 8318023 87 - 11907090 -------------------ACGCCCCGCCCCAGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUUGAACGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC -------------------.............(((..((..((((...((((((..------...))))))....))))..))(((((((.....)))))))....)))... ( -11.90, z-score = -1.16, R) >droMoj3.scaffold_6473 8863688 101 - 16943266 -----GCUCUCUCUCUCUCUCUUGGUCAUUCAGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUCGAGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC -----((((.............(((.(((((...)))))...)))...((((((..------...)))))).....))))((((((((((.....)))))))..)))..... ( -16.50, z-score = -2.54, R) >droGri2.scaffold_15203 10296754 105 + 11997470 -AUCCUCUCCCGUGGCAUCAUGCCAUCCACCAGUGAAUGUAUUCAUCAAUGCAACA------CAUUUGCAUAUUUUCAGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC -..........(((((.....)))))......(..(((((.((((....)).)).)------))))..).........((((((((((((.....)))))))..)))))... ( -21.30, z-score = -3.57, R) >consensus _A_CCACCACCUC__CCC___UCAGUGGGCCAGUGAAUGUAUUCAUCAAUGCAACA______CAUUUGCAUAUUUCGAGCGUAAAAAUUAAUCACUAAUUUUAAUACGCAAC ................................(((((....)))))..((((((...........)))))).......((((((((((((.....)))))))..)))))... (-12.14 = -12.32 + 0.18)

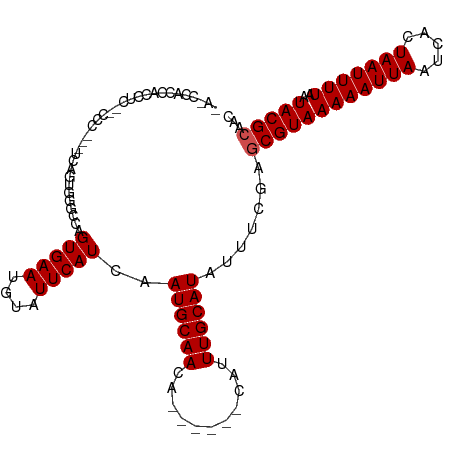

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:32 2011