
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,369,356 – 7,369,447 |
| Length | 91 |
| Max. P | 0.657764 |

| Location | 7,369,356 – 7,369,447 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.51 |
| Shannon entropy | 0.73100 |
| G+C content | 0.50301 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -11.68 |
| Energy contribution | -11.85 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
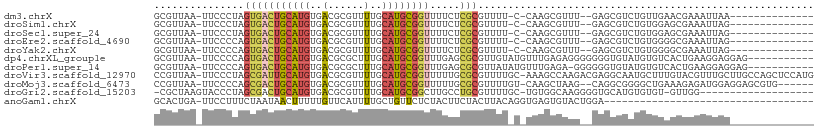
>dm3.chrX 7369356 91 - 22422827 GCGUUAA-UUCCCUAGUGACUGCAUGUGACGCGUUUUGCAUGCGGUUUUCUCGCGUUUU-C-CAAGCGUUU--GAGCGUCUGUUGAACGAAAUUAA-------------- (((....-......((.(((((((((..(......)..)))))))))..)))))((((.-.-((..(((..--..)))..))..))))........-------------- ( -26.20, z-score = -1.25, R) >droSim1.chrX 5896384 91 - 17042790 GCGUUAA-UUCCCUAGUGACUGCAUGUGACGCGUUUUGCAUGCGGUUUUCUCGCGUUUU-C-CAAGCGUUU--GAGCGUCUGUGGAGCGAAAUUAG-------------- .......-....((((((((((((((..(......)..)))))))))......(((((.-.-((..(((..--..)))..))..)))))..)))))-------------- ( -28.00, z-score = -1.12, R) >droSec1.super_24 578924 91 - 912625 GCGUUAA-UUCCCUAGUGACUGCAUGUGACGCGUUUUGCAUGCGGUUUUCUCGCGUUUU-C-CAAGCGUUU--GAGCGUCUGUGGAGCGAAAUUAG-------------- .......-....((((((((((((((..(......)..)))))))))......(((((.-.-((..(((..--..)))..))..)))))..)))))-------------- ( -28.00, z-score = -1.12, R) >droEre2.scaffold_4690 16281230 91 - 18748788 GCGUUAA-UUCCCCAGUGACUGCAUGUGACGCGUUUUGCAUGCGGUUUUCUCGCGUUUU-C-CAAGCGUUU--GAGCGUCUGUGGGGCGAAAUUAG-------------- ....(((-(((((((..(((((((((..(......)..))))))((((...(((.....-.-...)))...--)))))))..)))))...))))).-------------- ( -28.60, z-score = -0.89, R) >droYak2.chrX 7798218 91 + 21770863 GCGUUAA-UUCCCCAGUGACUGCAUGUGACGCGUUUUGCAUGCGGUUUUCUCGCGUUUU-C-CAAGCGUUU--GAGCGUCUGUGGGGCGAAAUUAG-------------- ....(((-(((((((..(((((((((..(......)..))))))((((...(((.....-.-...)))...--)))))))..)))))...))))).-------------- ( -28.60, z-score = -0.89, R) >dp4.chrXL_group1e 11355170 98 - 12523060 GCGUUAA-UUCCCCAGUGACUGCAUGUGACGCGCUUUGCAUGCGGUUUGAGCGCGUUGUAUGUUUGAGAGGGGGGGUGUAUGUGUCACUGAAGGAGGAG----------- ((((..(-((((((..(....(((((..((((((((.((.....))..))))))))..)))))....)..)))))))..))))................----------- ( -36.40, z-score = -2.31, R) >droPer1.super_14 801986 97 - 2168203 GCGUUAA-UUCCCCAGUGACUGCAUGUGACGCGCUUUGCAUGCGGUUUGAGCGCGUUAUAUGUUUGAGA-GGGGGGUGUAUGUGUCACUGAAGGAGGAG----------- ((((..(-((((((.......(((((((((((((((.((.....))..)))))))))))))))......-)))))))..))))................----------- ( -36.02, z-score = -2.54, R) >droVir3.scaffold_12970 3580251 108 - 11907090 CCGUUAA-UUCCCUAGCGAUUGCAUGUGACGCGUUUUGCAUGCGGUUUUUGCGCGUUUUGC-AAAGCCAAGACGAGGCAAUGCUUUGUACGUUUGCUUGCCAGCUCCAUG .(((((.-.....)))))....((((.((.((((((((((.(((((....)).)))..)))-)))))........(((((.((...........))))))).)))))))) ( -31.40, z-score = -0.08, R) >droMoj3.scaffold_6473 8071007 100 - 16943266 CCGUUAA-UUCCCCAGCGACUGCAUGUGACGCGUUUUGCAUGCGGUUUUUGCGCGUUUUGU-CAAGCUAAG--CAGGCGGGGCUGAAAGAGAUGGAGGAGCGUG------ .(((...-.(((((.(((((((((((..(......)..)))))))).....)))(((((..-..((((..(--....)..))))....))))))).))))))..------ ( -31.10, z-score = 0.57, R) >droGri2.scaffold_15203 1691071 88 + 11997470 -CGCUAAGUACCCUAGCGACUGCAUGUGACGCGUUUUGCAUGCGGCUUGCCUGCGUUUUGC-UGUGGCAAGGGGUGCAUGUGUGU-GUUGG------------------- -(((...((((((......(((((((..(......)..)))))))((((((.((.....))-...))))))))))))..)))...-.....------------------- ( -34.50, z-score = -0.92, R) >anoGam1.chrX 19864919 74 + 22145176 GCACUGA-UUCCUUUCUAAUAACUUUUUGUUCAUUUUGCUGUUCUCUACUUCUACUUACAGGUGAGUGUACUGGA----------------------------------- (((((.(-(...........(((.....))).......((((...............)))))).)))))......----------------------------------- ( -7.36, z-score = 1.04, R) >consensus GCGUUAA_UUCCCUAGUGACUGCAUGUGACGCGUUUUGCAUGCGGUUUUCUCGCGUUUU_C_CAAGCGUUG__GAGCGUCUGUGGAGCGAAAUUAG______________ ...............(((((((((((..(......)..)))))))).....)))........................................................ (-11.68 = -11.85 + 0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:31 2011