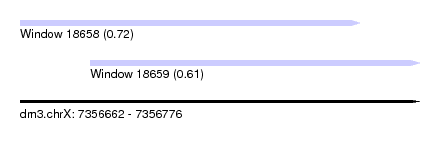
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,356,662 – 7,356,776 |
| Length | 114 |
| Max. P | 0.721090 |

| Location | 7,356,662 – 7,356,759 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.85 |
| Shannon entropy | 0.30901 |
| G+C content | 0.34414 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -14.59 |
| Energy contribution | -15.81 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
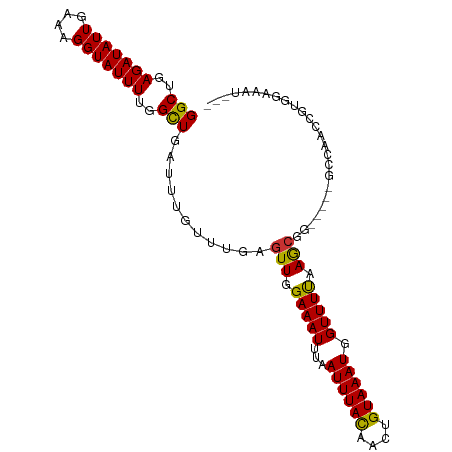
>dm3.chrX 7356662 97 + 22422827 --UGAAAACAAAAAAUG--CCAGGGGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUG-GUUUUAAGC --..............(--((...(((..(((((((....)))))))..))).....((((((((((.............))))).))))))-))....... ( -20.72, z-score = -2.12, R) >droSim1.chrX 5883284 97 + 17042790 --UGAAAACAAAAAAUG--CCAGGGGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUG-GUUUUAAGC --..............(--((...(((..(((((((....)))))))..))).....((((((((((.............))))).))))))-))....... ( -20.72, z-score = -2.12, R) >droSec1.super_24 565724 97 + 912625 --UGAAAACAAAAAAUG--CCAGGGGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUG-GUUUUAAGC --..............(--((...(((..(((((((....)))))))..))).....((((((((((.............))))).))))))-))....... ( -20.72, z-score = -2.12, R) >droAna3.scaffold_13117 205211 100 + 5790199 CAGGAACAGGGACAACAA-CCCCGGGCGGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUUCGACUGUAAAUG-GUUUUAAGC ..(((((.(((.......-.))).(((.((((((((....)))))))).))).....(((((((((((((((...)))))))))).))))).-))))).... ( -26.90, z-score = -2.44, R) >dp4.chrXL_group1e 11339854 99 + 12523060 -AACGGGACAGAAAAGAG-CCCUGGGCGAAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUG-GUUUUAAGC -..((((.(........)-.))))(((.((((((((....)))))))).)))...(..(((((((((.............))))).))))..-)........ ( -21.52, z-score = -1.58, R) >droPer1.super_14 784864 99 + 2168203 -AACGGGACAGAAAAGAG-CCCUGGGCGAAGAUAUUGAAAGGUAUUUUGGUUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUG-GUUUUAAGC -..((((.(........)-.))))(((.((((((((....)))))))).)))...(..(((((((((.............))))).))))..-)........ ( -19.02, z-score = -0.94, R) >droWil1.scaffold_180777 3429978 98 + 4753960 ---UGAGACGACAAAUAUGCCCAAGGCUAAGAUAUUGAAAGGUAUUUUCGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUAUAACUGUAAAUG-GUUUCAAAG ---((((((..((((((.......(((.((((((((....)))))))).)))....))))))(((((.((((...)))).))))).......-))))))... ( -20.70, z-score = -1.73, R) >droMoj3.scaffold_6473 8046269 97 + 16943266 -----CGAUGGCAAAUGCGCCUGGGGCGCAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUAUAACUGUAAAUGUAUUUUAGAC -----((((.(((((((((((...)))))(((((((....)))))))......))))))....))))(((((...((((((....))))))..))))).... ( -20.50, z-score = -1.18, R) >consensus __UGAAGACAAAAAAUA__CCCGGGGCGGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUG_GUUUUAAGC ....(((((...............(((.((((((((....)))))))).))).....((((((((((.((((...)))).))))).)))))..))))).... (-14.59 = -15.81 + 1.22)


| Location | 7,356,682 – 7,356,776 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 88.13 |
| Shannon entropy | 0.22990 |
| G+C content | 0.36196 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -15.31 |
| Energy contribution | -17.00 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7356682 94 + 22422827 GGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGG-----GCCAACCGUGGAAAU--- (((..(((((((....)))))))..)))...(..(((((((((.............))))).))))..).(((..((((-----.....))))..))).--- ( -25.02, z-score = -2.82, R) >droSim1.chrX 5883304 94 + 17042790 GGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGG-----GCCAACCGUGGAAAU--- (((..(((((((....)))))))..)))...(..(((((((((.............))))).))))..).(((..((((-----.....))))..))).--- ( -25.02, z-score = -2.82, R) >droSec1.super_24 565744 94 + 912625 GGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGG-----GCCAACCGUGGAAAU--- (((..(((((((....)))))))..)))...(..(((((((((.............))))).))))..).(((..((((-----.....))))..))).--- ( -25.02, z-score = -2.82, R) >droYak2.chrX 7785198 99 - 21770863 GGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGGCGAGUACCAACCGUGGAAAU--- (((..(((((((....)))))))..))).((((((....(((.(((((...((((((....)))))).))))).))).)))))).(((....)))....--- ( -24.70, z-score = -2.34, R) >droEre2.scaffold_4690 16268522 94 + 18748788 GGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGC-----ACCAGCCGUGGAAAU--- ((((((((((((....)))))))(((((...(..(((((((((.............))))).))))..).....))).)-----).)))))........--- ( -22.92, z-score = -1.79, R) >dp4.chrXL_group1e 11339876 94 + 12523060 GGCGAAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGU-----UGCGCUCACGGAGCG--- (((.((((((((....)))))))).))).....((....(((.(((((...((((((....)))))).))))).)))..-----.))((((...)))).--- ( -23.30, z-score = -1.73, R) >droPer1.super_14 784886 94 + 2168203 GGCGAAGAUAUUGAAAGGUAUUUUGGUUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGU-----UGCGCUCACGGAGCG--- (((.((((((((....)))))))).))).....((....(((.(((((...((((((....)))))).))))).)))..-----.))((((...)))).--- ( -20.80, z-score = -1.12, R) >droWil1.scaffold_180777 3429999 97 + 4753960 GGCUAAGAUAUUGAAAGGUAUUUUCGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUAUAACUGUAAAUGGUUUCAAAGCA-----GCUAAACAGGGUGAGAGU (((.((((((((....)))))))).)))..(((((((.((((((((((...((((((....)))))).)))))....))-----))))))))))........ ( -21.00, z-score = -1.60, R) >consensus GGCUGAGAUAUUGAAAGGUAUUUUGGCUGAUUUGUUUGAGUUGGAAAUUUAAUUUACAACUGUAAAUGGUUUUAAGCGG_____GCCAACCGUGGAAAU___ (((..(((((((....)))))))..)))...........(((.(((((...((((((....)))))).))))).)))......................... (-15.31 = -17.00 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:31 2011