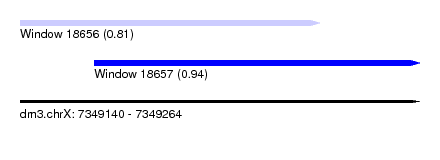
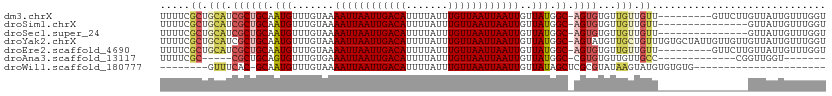
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,349,140 – 7,349,264 |
| Length | 124 |
| Max. P | 0.936761 |

| Location | 7,349,140 – 7,349,233 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Shannon entropy | 0.42125 |
| G+C content | 0.31480 |
| Mean single sequence MFE | -20.89 |
| Consensus MFE | -11.18 |
| Energy contribution | -10.99 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810466 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
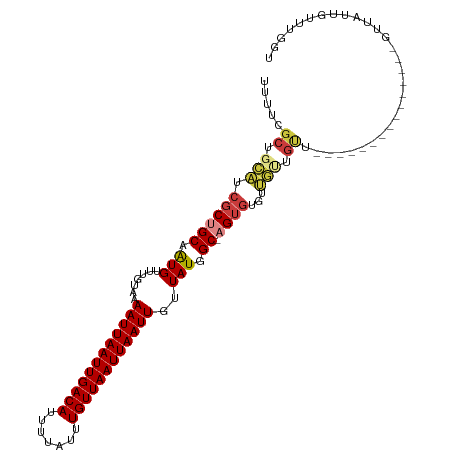
>dm3.chrX 7349140 93 + 22422827 ------UGUGUGUUUUUUUUAACCGUGUAUUUUCGC-UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAG ------................((((((((...((.-((((....)))).))...))).((((((((((((.......))))))))))))..)))))... ( -17.00, z-score = -0.70, R) >droSim1.chrX 5875260 93 + 17042790 ------UGUGUGUUUUUUUUAACCGUGUAUUUUCGC-UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAG ------................((((((((...((.-((((....)))).))...))).((((((((((((.......))))))))))))..)))))... ( -17.00, z-score = -0.70, R) >droSec1.super_24 558364 93 + 912625 ------UGUGUGUUUUUUUUAACCGUGUAUUUUCGC-UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAG ------................((((((((...((.-((((....)))).))...))).((((((((((((.......))))))))))))..)))))... ( -17.00, z-score = -0.70, R) >droYak2.chrX 7776777 93 - 21770863 ------UGUGUGUUUUUUUUAAAUGUGUAUUUUCGC-UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAG ------((..(((((.....)))))..)).....((-((((....))).......((((((((((((((((.......)))))))))))).))))))).. ( -16.50, z-score = -0.47, R) >droEre2.scaffold_4690 16260974 93 + 18748788 ------UGUGUGUUUUUUUUAACCGUGUAUUUUCGC-UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAG ------................((((((((...((.-((((....)))).))...))).((((((((((((.......))))))))))))..)))))... ( -17.00, z-score = -0.70, R) >droAna3.scaffold_13117 194269 78 + 5790199 -----------GCUUUUUUCGCCU-----UUUUCGC------CGCUGCAGUGUUUGUGAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCCG -----------.........(((.-----...((((------(((....)))...))))((((((((((((.......)))))))))))).....))).. ( -17.40, z-score = -2.61, R) >dp4.chrXL_group1e 11330895 98 + 12523060 UUUAUUUUUUCGCAUUGGUGUACGGGCCGCCCGCCCGUACGCUGCUGCUGUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUAGC-- ...........(((..(((((((((((.....)))))))))))..))).....((((((((((((((((((.......)))))))))))).)))))).-- ( -35.90, z-score = -5.70, R) >droPer1.super_14 775795 96 + 2168203 UUUAUUUUUUCGCUUUGGUGUACGGGCGGGCGGC--AUACGCUGCCGCUGUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUAGC-- ...........(((.(((..((((((((((((((--(.....))))))).)))))))).((((((((((((.......)))))))))))).))).)))-- ( -29.30, z-score = -3.41, R) >consensus ______UGUGUGUUUUUUUUAACCGUGUAUUUUCGC_UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAG ...................................................(((.....((((((((((((.......)))))))))))).....))).. (-11.18 = -10.99 + -0.19)
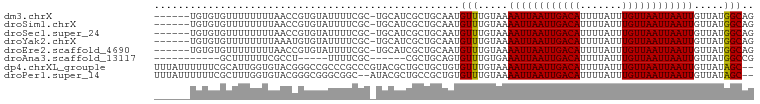
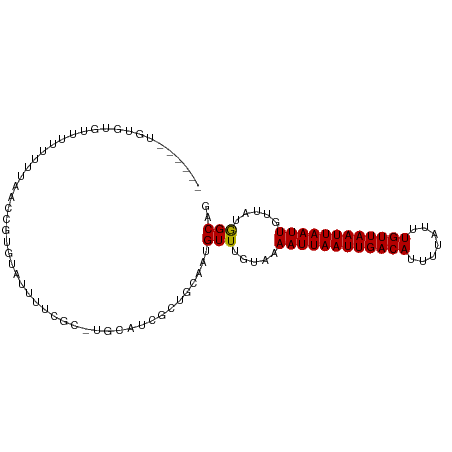
| Location | 7,349,163 – 7,349,264 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Shannon entropy | 0.40362 |
| G+C content | 0.30500 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.71 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936761 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 7349163 101 + 22422827 UUUUCGCUGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGC-AGUGUGUUGUUGUU---------GUUCUUGUUAUUGUUUGGU .....((.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))-))))))).))....---------................... ( -21.20, z-score = -1.86, R) >droSim1.chrX 5875283 95 + 17042790 UUUUCGCUGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGC-AGUGUGUUGUUGUU---------------GUUAUUGUUUGGU .....((.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))-))))))).))....---------------............. ( -21.20, z-score = -1.84, R) >droSec1.super_24 558387 95 + 912625 UUUUCGCUGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGC-AGUGUGUUGUUGUU---------------GUUAUUGUUUGGU .....((.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))-))))))).))....---------------............. ( -21.20, z-score = -1.84, R) >droYak2.chrX 7776800 110 - 21770863 UUUUCGCUGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGC-AGUAUGUUGCUGUUUGUGCUAUUGUUGUUGUUAUUGUUUGGU ........((((.((.(((((((((((..((((((((((((.......))))))))))))......))-)).))))))).))..))))....................... ( -23.50, z-score = -1.38, R) >droEre2.scaffold_4690 16260997 101 + 18748788 UUUUCGCUGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGC-AGUGUGUUGUUGUU---------GUUCUUGUUAUUGUUUGGU .....((.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))-))))))).))....---------................... ( -21.20, z-score = -1.86, R) >droAna3.scaffold_13117 194282 85 + 5790199 UUUUCGC-----CGCUGCAGUGUUUGUGAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGC-CGUGUGUUGUUGCC-------------CGGUUGGU------- ...((((-----(((....)))...))))((((((((((((.......)))))))))))).....(((-((.((......)).-------------)))))...------- ( -19.40, z-score = -1.26, R) >droWil1.scaffold_180777 3436269 80 + 4753960 --------GUUUCAC-GCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUAGCUCGCGUAUAAGUAUGUGUGUG---------------------- --------....(((-((.....((((((((((((((((((.......)))))))))))).))))))...(((((....))))))))))---------------------- ( -18.90, z-score = -2.41, R) >consensus UUUUCGCUGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGC_AGUGUGUUGUUGUU_______________GUUAUUGUUUGGU .....((.(((.(((.((...(((.....((((((((((((.......)))))))))))).....)))..))))).))).))............................. (-14.52 = -14.71 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:29 2011