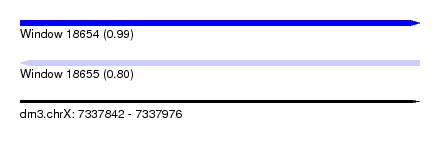
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,337,842 – 7,337,976 |
| Length | 134 |
| Max. P | 0.988343 |

| Location | 7,337,842 – 7,337,976 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 157 |
| Reading direction | forward |
| Mean pairwise identity | 68.10 |
| Shannon entropy | 0.57215 |
| G+C content | 0.44280 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -16.57 |
| Energy contribution | -16.99 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
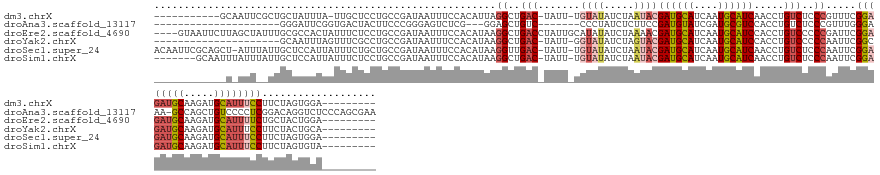
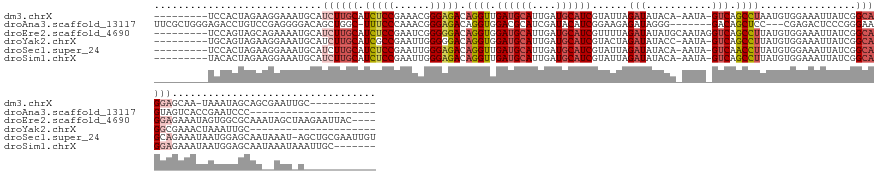
>dm3.chrX 7337842 134 + 22422827 -----------GCAAUUCGCUGCUAUUUA-UUGCUCCUGCCGAUAAUUUCCACAUUAGGCUGAC-UAUU-UGUAUAUCUAAUACGAUGCAUCAAUGCAUCAACCUGUCUCCCGUUUCGGAGAUGCAAGAUGCAUUUCCUUCUAGUGGA--------- -----------(((......)))...(((-((((....).))))))..(((((....((..(((-....-.((((.....))))((((((....)))))).....)))..)).....((((((((.....)))))))).....)))))--------- ( -38.10, z-score = -3.28, R) >droAna3.scaffold_13117 184564 125 + 5790199 ---------------------GGGAUUCGGUGACUACUUCCCGGGAGUCUCG---GGAGCUGUC-------CCCUAUCUCUUCCGAUGUAUCGAUGCGUCCACCUGUCUCCCGUUUGGGAAA-GCCAGCUGUCCCCUCGGACAGGUCUCCCAGCGAA ---------------------((((...((.(((...((((((((...))))---))))..)))-------.))..........((((((....)))))).((((((...(((...((((.(-(....)).))))..))))))))).))))...... ( -45.30, z-score = -0.44, R) >droEre2.scaffold_4690 16249336 144 + 18748788 ----GUAAUUCUUAGCUAUUUGCGCCACUAUUUCUCCUGCCGAUAAUUUCCACAUAAGGCUGACCUAUUGCAUAUAUCUAAAACGAUGCAUCAAUGCAUCCACCUGUCCCCCGAUUCGGAGAUGCAAGAUGCAUUUUCUGCUACUGGA--------- ----(((....((((.(((.((((....((..((....(((................))).))..)).)))).))).))))...((((((((..((((((..(..(((....)))..)..)))))).))))))))...))).......--------- ( -28.39, z-score = -0.41, R) >droYak2.chrX 7764441 125 - 21770863 ---------------------GCAAUUUAGUUUCGCCUGCCGAUAAUUUCCACAUAAGGCUGAC-UAUU-GGUAUAUCUAGUACGAUGCAUCAAUGCAUCCACCUGUCCCCCAAUUCGGCGAUGCAAGAUGCAUUUCCUUCUACUGCA--------- ---------------------(((.(((.((.(((((((((((((.((.((......))..)).-))))-))))....(((...((((((....))))))...)))...........))))).)).))))))................--------- ( -30.60, z-score = -1.85, R) >droSec1.super_24 547099 145 + 912625 ACAAUUCGCAGCU-AUUUAUUGCUCCAUUAUUUCUGCUGCCGAUAAUUUCCACAUAAGGUUGAC-UAUU-UGUAUAUCUAAUACGAUGCAUCAAUGCAUCAACCUGUCUCCCAAUUCGGAGAUGCAAGAUGCAUUUCCUUCUAGUGGA--------- ...(((.(((((.-.....................))))).)))....(((((....((..(((-....-.((((.....))))((((((....)))))).....)))..)).....((((((((.....)))))))).....)))))--------- ( -41.65, z-score = -4.20, R) >droSim1.chrX 5865064 139 + 17042790 -------GCAAUUUAUUUAUUGCUCCAUUAUUUCUCCUGCCGAUAAUUUCCACAUAAGGCUGAC-UAUU-UGUAUAUCUAAUACGAUGCAUCAAUGCAUCAACCUGUCUCCCAAUUCGGAGAUGCAAGAUGCAUUUCCUUCUAGUGUA--------- -------(((((......)))))...........................(((....((..(((-....-.((((.....))))((((((....)))))).....)))..)).....((((((((.....)))))))).....)))..--------- ( -33.60, z-score = -3.47, R) >consensus ____________U_AUU__UUGCUACAUUAUUUCUCCUGCCGAUAAUUUCCACAUAAGGCUGAC_UAUU_UGUAUAUCUAAUACGAUGCAUCAAUGCAUCAACCUGUCUCCCAAUUCGGAGAUGCAAGAUGCAUUUCCUUCUAGUGGA_________ ....................................(.(((................))).)......................((((((....)))))).................((((((((.....))))))))................... (-16.57 = -16.99 + 0.42)
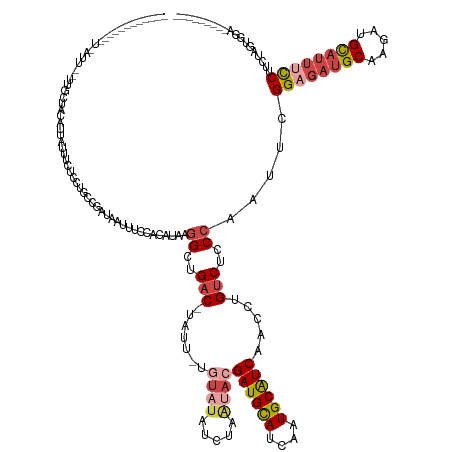
| Location | 7,337,842 – 7,337,976 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 157 |
| Reading direction | reverse |
| Mean pairwise identity | 68.10 |
| Shannon entropy | 0.57215 |
| G+C content | 0.44280 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -16.98 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
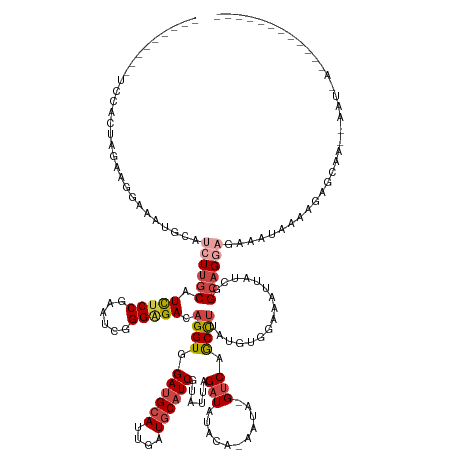
>dm3.chrX 7337842 134 - 22422827 ---------UCCACUAGAAGGAAAUGCAUCUUGCAUCUCCGAAACGGGAGACAGGUUGAUGCAUUGAUGCAUCGUAUUAGAUAUACA-AAUA-GUCAGCCUAAUGUGGAAAUUAUCGGCAGGAGCAA-UAAAUAGCAGCGAAUUGC----------- ---------(((.......)))..(((.((((((.(((((......))))).(((((((((((....)))...((((.....)))).-....-))))))))................))))))))).-......((((....))))----------- ( -35.70, z-score = -1.66, R) >droAna3.scaffold_13117 184564 125 - 5790199 UUCGCUGGGAGACCUGUCCGAGGGGACAGCUGGC-UUUCCCAAACGGGAGACAGGUGGACGCAUCGAUACAUCGGAAGAGAUAGGG-------GACAGCUCC---CGAGACUCCCGGGAAGUAGUCACCGAAUCCC--------------------- ......((((..((((((((((.(..((.(((..-((((((....))))))))).))..).).))).....((....)))))))).-------(((.(((((---((.......)))).))).)))......))))--------------------- ( -48.60, z-score = -1.03, R) >droEre2.scaffold_4690 16249336 144 - 18748788 ---------UCCAGUAGCAGAAAAUGCAUCUUGCAUCUCCGAAUCGGGGGACAGGUGGAUGCAUUGAUGCAUCGUUUUAGAUAUAUGCAAUAGGUCAGCCUUAUGUGGAAAUUAUCGGCAGGAGAAAUAGUGGCGCAAAUAGCUAAGAAUUAC---- ---------.......(((...(((((((((....(((((......))))).....)))))))))..)))....((((((.(((.(((...(((....)))..(((.((.....)).)))..............))).))).)))))).....---- ( -37.30, z-score = -0.56, R) >droYak2.chrX 7764441 125 + 21770863 ---------UGCAGUAGAAGGAAAUGCAUCUUGCAUCGCCGAAUUGGGGGACAGGUGGAUGCAUUGAUGCAUCGUACUAGAUAUACC-AAUA-GUCAGCCUUAUGUGGAAAUUAUCGGCAGGCGAAACUAAAUUGC--------------------- ---------.(((((....((....(((((.(((((((((......(....).))).))))))..)))))(((......)))...))-..((-((..((((...((.((.....)).))))))...)))).)))))--------------------- ( -36.90, z-score = -1.74, R) >droSec1.super_24 547099 145 - 912625 ---------UCCACUAGAAGGAAAUGCAUCUUGCAUCUCCGAAUUGGGAGACAGGUUGAUGCAUUGAUGCAUCGUAUUAGAUAUACA-AAUA-GUCAACCUUAUGUGGAAAUUAUCGGCAGCAGAAAUAAUGGAGCAAUAAAU-AGCUGCGAAUUGU ---------(((((...((((..(((((((.((((((.((......(....).))..))))))..))))))).((((.....)))).-....-.....))))..)))))........(((((.....................-.)))))....... ( -41.65, z-score = -2.64, R) >droSim1.chrX 5865064 139 - 17042790 ---------UACACUAGAAGGAAAUGCAUCUUGCAUCUCCGAAUUGGGAGACAGGUUGAUGCAUUGAUGCAUCGUAUUAGAUAUACA-AAUA-GUCAGCCUUAUGUGGAAAUUAUCGGCAGGAGAAAUAAUGGAGCAAUAAAUAAAUUGC------- ---------..(((...((((..(((((((.((((((.((......(....).))..))))))..))))))).((((.....)))).-....-.....))))..)))...........((..(....)..))..(((((......)))))------- ( -33.70, z-score = -1.65, R) >consensus _________UCCACUAGAAGGAAAUGCAUCUUGCAUCUCCGAAUCGGGAGACAGGUGGAUGCAUUGAUGCAUCGUAUUAGAUAUACA_AAUA_GUCAGCCUUAUGUGGAAAUUAUCGGCAGGAGAAAUAAAAGAGCAA__AAU_A____________ ......................((((((((..((.(((((......)))))...)).))))))))..((((((......))).................((..(((.((.....)).)))..))..........))).................... (-16.98 = -18.10 + 1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:27 2011