
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,505,648 – 10,505,779 |
| Length | 131 |
| Max. P | 0.738261 |

| Location | 10,505,648 – 10,505,747 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.37 |
| Shannon entropy | 0.52929 |
| G+C content | 0.47822 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -16.09 |
| Energy contribution | -17.27 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
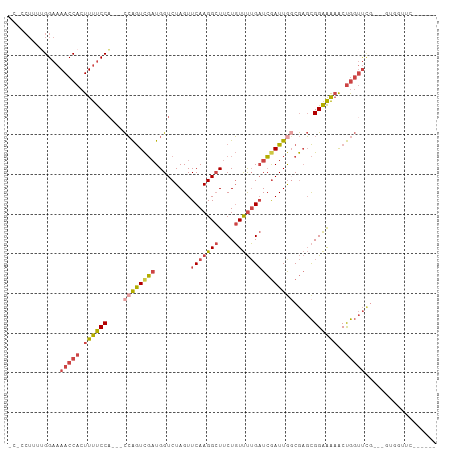
>dm3.chr2L 10505648 99 - 23011544 GCGCCUUUGGGAAAACCACUUUUCCU---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUAGCGAGCGGAAAAACUGGUUUGACCGUAGUUC------ ....((.(((..((((((.((((..(---((..(((.(((((....(((((((....)))))))..))))).)))..))))))).))))))..))).))...------ ( -28.90, z-score = -1.19, R) >droAna3.scaffold_12916 14351270 102 - 16180835 -----UCUCGGAAAACCACUUCCCCACCAUGGAUUUCUGGUCUAGUUCAAGG-UUCUUUCCAGCUAGAUUGCCACCUGGGGGCGCCACACAGAAACUGGUUGCGGUUC -----........((((...((((((...(((......(((((((((...((-......))))))))))).)))..)))))).((.((.(((...))))).)))))). ( -27.90, z-score = 0.41, R) >droEre2.scaffold_4929 11719103 98 + 26641161 -AGCCUGCUGGAAAACCACUUUUCCA---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGGCGAGCGGAAAAACUGGUUCGCCUGUGGUUC------ -((((.((.((..(((((.((((((.---(..((((((.(......(((((((....))))))).).))))))..).))))))..)))))..)).)))))).------ ( -32.30, z-score = -1.66, R) >droYak2.chr2L 6913224 91 - 22324452 --------UGGAAAACCACUUUUCCA---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGCCGAGCGGAAAAACUGGUUCGCUUGUGCUUC------ --------(((((((....)))))))---..((((((.........(((((((....)))))))))))))((((((((((........)))))))).))...------ ( -26.60, z-score = -1.37, R) >droSec1.super_3 5932587 89 - 7220098 ACACCUUUUGGAAAACCACUUUUCCA--CCCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCAAUUGGCGAGCGGAAAAACUGGUUC----------------- ...((....))..(((((.((((((.--(...((((((........(((((((....)))))))...))))))..).))))))..))))).----------------- ( -22.10, z-score = -0.49, R) >droSim1.chr2L 10310564 88 - 22036055 ACUACUUUUGGAAAACCACUUUUCCA---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGGCGAGCGGAAAAACUGGUUC----------------- .............(((((.((((((.---(..((((((.(......(((((((....))))))).).))))))..).))))))..))))).----------------- ( -23.50, z-score = -1.13, R) >consensus _C_CCUUUUGGAAAACCACUUUUCCA___CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGGCGAGCGGAAAAACUGGUUCG___GUGGUUC______ .............(((((.((((((....((((((((.........(((((((....))))))))))))))).....))))))..))))).................. (-16.09 = -17.27 + 1.17)

| Location | 10,505,671 – 10,505,779 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.96 |
| Shannon entropy | 0.52748 |
| G+C content | 0.52536 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -13.17 |
| Energy contribution | -14.98 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
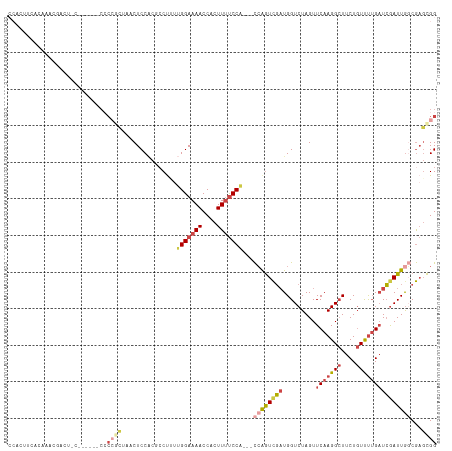
>dm3.chr2L 10505671 108 - 23011544 CCACAUCACAAACGACUCC------CCCCGCGAACUCCGCGCCUUUGGGAAAACCACUUUUCCU---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUAGCGAGCGG ((.(.((.(...(((((..------...((((.....)))).....(((((((....)))))))---..))))).(((((....(((((((....)))))))..)))))).))).)) ( -26.10, z-score = -0.27, R) >droAna3.scaffold_12916 14351299 91 - 16180835 ------------------------GCACAAGUCACUCCU-UCCUCUCGGAAAACCACUUCCCCACCAUGGAUUUCUGGUCUAGUUCAAGG-UUCUUUCCAGCUAGAUUGCCACCUGG ------------------------(((............-......((((((.(((...........))).))))))((((((((...((-......)))))))))))))....... ( -16.90, z-score = 0.20, R) >droEre2.scaffold_4929 11719126 113 + 26641161 CCACUCCACAACCCACCACCCGCCGCCCCCCGAACUCCA-GCCUGCUGGAAAACCACUUUUCCA---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGGCGAGCGG ...................((((((((...(((...(((-..(((.(((((((....)))))))---.)))....)))......(((((((....))))))))))...)))).)))) ( -34.20, z-score = -2.87, R) >droYak2.chr2L 6913247 83 - 22324452 -------------------------------CCACUCCACCCCCCUUGGAAAACCACUUUUCCA---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGCCGAGCGG -------------------------------((.(((.........(((((((....)))))))---.(((((((.........(((((((....))))))))))))))..))).)) ( -21.60, z-score = -1.49, R) >droSec1.super_3 5932599 109 - 7220098 CCACUUCACAAACGACUUC------CCCCGCUAACUCCACACCUUUUGGAAAACCACUUUUCCA--CCCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCAAUUGGCGAGCGG ...................------..(((((..............(((((((....)))))))--....((((((........(((((((....)))))))...)))))).))))) ( -23.20, z-score = -0.97, R) >droSim1.chr2L 10310576 108 - 22036055 CCACUUCACAAACGACUCC------CCCCGCUAACUCCACUACUUUUGGAAAACCACUUUUCCA---CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGGCGAGCGG ...................------..(((((..............(((((((....)))))))---((((((((.........(((((((....)))))))))))))))..))))) ( -27.90, z-score = -2.14, R) >consensus CCACUUCACAAACGACU_C______CCCCGCUAACUCCACGCCUUUUGGAAAACCACUUUUCCA___CCAGUCGAUGGUCUAGUUCAAGGCUUCUGUUUUGAUCGAUUGGCGAGCGG ...........................((((................((((((....))))))....((((((((.........(((((((....)))))))))))))))...)))) (-13.17 = -14.98 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:23 2011