
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,333,680 – 7,333,779 |
| Length | 99 |
| Max. P | 0.876749 |

| Location | 7,333,680 – 7,333,779 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Shannon entropy | 0.45961 |
| G+C content | 0.50805 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.81 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
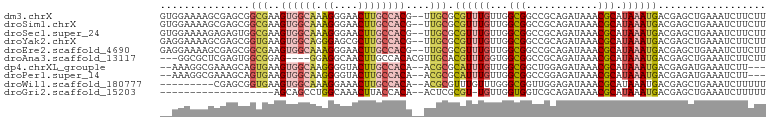
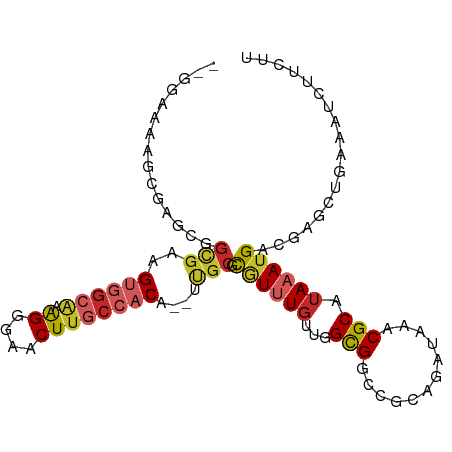
>dm3.chrX 7333680 99 - 22422827 GUGGAAAAGCGAGCGGCGAAGUGGCAAAGGGAACUUGCCACG--UUGCGCGUUUGUUGGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUU (.(((..(((...((((((.(((((((.(....)))))))).--))))(((((((((.((....)).)))))))))........)).)))....))).).. ( -37.40, z-score = -2.31, R) >droSim1.chrX 5860802 99 - 17042790 GUGGAAAAGCGAGCGGCGAAGUGGCAAAGGGAACUUGCCACG--UUGCGCGUUUGUUGGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUU (.(((..(((...((((((.(((((((.(....)))))))).--))))(((((((((.((....)).)))))))))........)).)))....))).).. ( -37.40, z-score = -2.31, R) >droSec1.super_24 542805 99 - 912625 GUGGAAAAGAGAGUGGCGAAGUGGCAAAGGGAACUUGCCACG--UUGCGCGUUUGUUGGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUU ......((((.(((.((((.(((((((.(....)))))))).--))))(((((((((.((....)).)))))))))...........)))....))))... ( -33.70, z-score = -2.01, R) >droYak2.chrX 7760814 99 + 21770863 GAGGAAAAGCGAGCGGUGAAGUGGCAGGGAGCGCUUGCCACG--UUGCGCGUUUGUUGGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUU (((((..(((...(((..(.((((((((.....)))))))).--)..)(((((((((.((....)).)))))))))........)).)))....))))).. ( -40.30, z-score = -2.65, R) >droEre2.scaffold_4690 16244775 99 - 18748788 GAGGAAAAGCGAGCGGCGAAGUGGCAAAGGGAACUUGCCACG--UUGCGCGUUUGUUGGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUU (((((..(((...((((((.(((((((.(....)))))))).--))))(((((((((.((....)).)))))))))........)).)))....))))).. ( -41.50, z-score = -3.84, R) >droAna3.scaffold_13117 181545 94 - 5790199 ---GGCGCUCGAGUGGCGGAG----GGAGGCAACUUGCCACACGUUGCACGUUUGGUGGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUU ---...(((((.(..((((.(----((((....))).)).).)))..).((((((...((....))...)))))).........)))))............ ( -29.90, z-score = -0.14, R) >dp4.chrXL_group1e 11313214 94 - 12523060 --AAAGGCGAAAGCAGUGAAGUGGCAAGGGGUACUUGCCACA--ACGCGCAUUUGUUGGCGGCUGGAGAUAAACGCAUAAAUGACGAGAUGAAAUCUU--- --....((....)).(((..((((((((.....)))))))).--.)))((((((((..(((..((....))..)))))))))).)(((((...)))))--- ( -31.20, z-score = -3.25, R) >droPer1.super_14 756604 94 - 2168203 --AAAGGCGAAAGCAGUGAAGUGGCAAGGGGUACUUGCCACA--ACGCGCAUUUGUUGGCGGCCGGAGAUAAACGCAUAAAUGACGAGAUGAAAUCUU--- --....((....)).(((..((((((((.....)))))))).--.)))((((((((..(((............)))))))))).)(((((...)))))--- ( -30.60, z-score = -2.70, R) >droWil1.scaffold_180777 1302673 90 + 4753960 ---------CGAGCGGUGAAGUGGCAAAGGAAACUUGCCACA--ACGCGUUUGUUUGGGCGGUUGGAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUUUU ---------.((.((((...(((((((.(....)))))))).--..((((((((((..........))))))))))...........))))...))..... ( -28.90, z-score = -2.63, R) >droGri2.scaffold_15203 1642333 79 + 11997470 -------------------AGCAGCCUGGCAAACUUACCACA--ACUCGCGU-UGUUGGUGGUCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUUUU -------------------..((((.((.((...((((((((--((....))-)).))))))..((........)).....)).)).)))).......... ( -16.90, z-score = 0.35, R) >consensus __GGAAAAGCGAGCGGCGAAGUGGCAAAGGGAACUUGCCACA__UUGCGCGUUUGUUGGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUU ....................((((((.((....))))))))......((((((((...(((............))).)))))).))............... (-15.15 = -15.81 + 0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:26 2011