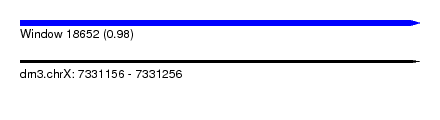
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,331,156 – 7,331,256 |
| Length | 100 |
| Max. P | 0.980591 |

| Location | 7,331,156 – 7,331,256 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 68.64 |
| Shannon entropy | 0.60509 |
| G+C content | 0.52621 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
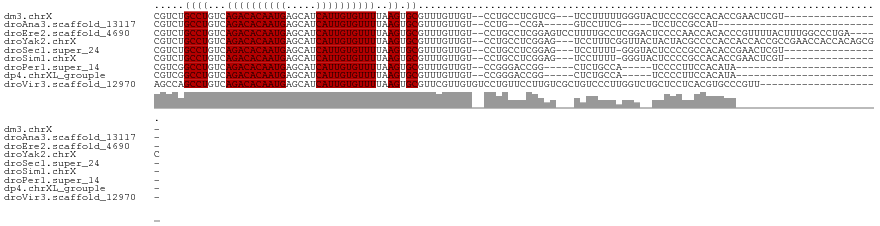
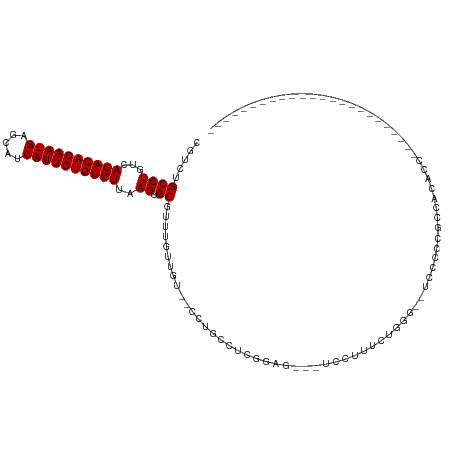
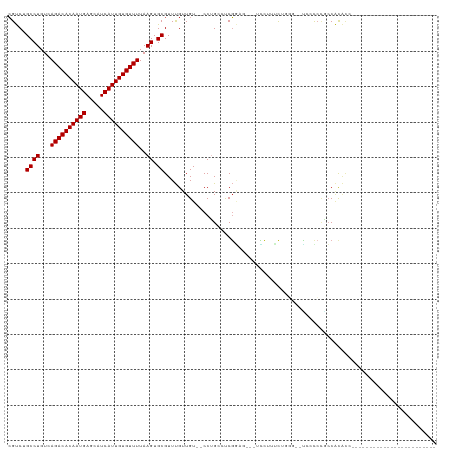
>dm3.chrX 7331156 100 + 22422827 CGUCUGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCUGCCUCGUCG---UCCUUUUUGGGUACUCCCCGCCACACCGAACUCGU---------------- .....((((...((((((((((.....))))))))))..)).))(((((.(((--...((...(..(---((((....))).))..)..)).))).)))))....---------------- ( -24.60, z-score = -1.06, R) >droAna3.scaffold_13117 179361 80 + 5790199 CGUCUGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCUG--CCGA-----GUCCUUCG-----UCCUCCGCCAU--------------------------- ((..(((((...((((((((((.....))))))))))..)).)))..))....--...(--(.((-----(..(...)-----..))).))...--------------------------- ( -19.10, z-score = -1.76, R) >droEre2.scaffold_4690 16242309 114 + 18748788 CGUCUGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCUGCCUCGGAGUCCUUUUGCCUCGGACUCCCCAACCACACCCGUUUUACUUUGGCCCUGA----- .....(((....((((((((((.....)))))))))).(((((((..(((.((--..((....(((((((.........))))))).)))).)))..)))...)))).))).....----- ( -31.30, z-score = -2.08, R) >droYak2.chrX 7758213 116 - 21770863 CGUCUGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCUGCCUCGGAG---UCCUUUCGGUUACUACUACGCCCCACCACCACCGCCGAACCACCACAGCGC .....((((...((((((((((.....))))))))))..)).))....(((((--..((..((((..---......((((....................))))))))..))..))))).. ( -26.35, z-score = 0.02, R) >droSec1.super_24 540306 99 + 912625 CGUCUGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCUGCCUCGGAG---UCCUUUU-GGGUACUCCCCGCCACACCGAACUCGU---------------- .....((((...((((((((((.....))))))))))..)).))(((((.(((--...((...((((---(((....-.)).)))))..)).))).)))))....---------------- ( -31.50, z-score = -2.53, R) >droSim1.chrX 5858303 99 + 17042790 CGUCUGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCUGCCUCGGAG---UCCUUUU-GGGUACUCCCCGCCACACCGAACUCGU---------------- .....((((...((((((((((.....))))))))))..)).))(((((.(((--...((...((((---(((....-.)).)))))..)).))).)))))....---------------- ( -31.50, z-score = -2.53, R) >droPer1.super_14 753657 85 + 2168203 CGUCGGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCGGGACCGG-----CUCUGCCA-----UCCCCUUCCACAUA------------------------ .....((((...((((((((((.....))))))))))..)).))...(((.(.--..((((..((-----(...))).-----))))...).)))..------------------------ ( -24.90, z-score = -1.27, R) >dp4.chrXL_group1e 11310357 85 + 12523060 CGUCGGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU--CCGGGACCGG-----CUCUGCCA-----UCCCCUUCCACAUA------------------------ .....((((...((((((((((.....))))))))))..)).))...(((.(.--..((((..((-----(...))).-----))))...).)))..------------------------ ( -24.90, z-score = -1.27, R) >droVir3.scaffold_12970 3526509 101 + 11907090 AGCCAGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUCGUUGUGUCCUGUUCCUUGUCGCUGUCCCUUGGUCUGCUCCUCACGUGCCCGUU-------------------- (((..((.(((((((((((((((((..((((........)))).))))))))))).))).........((.(.((...)).).)).....))).))..)))-------------------- ( -24.20, z-score = -1.12, R) >consensus CGUCUGCCUGUCAGACACAAUGAGCAUCAUUGUGUUUUAAGUGCGUUUGUUGU__CCUGCCUCGGAG___UCCUUUCUGGG__UCCCCCGCCACACC________________________ .....((((...((((((((((.....))))))))))..)).))............................................................................. (-15.22 = -15.22 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:25 2011