| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,326,609 – 7,326,707 |
| Length | 98 |
| Max. P | 0.700722 |

| Location | 7,326,609 – 7,326,707 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 62.46 |
| Shannon entropy | 0.78709 |
| G+C content | 0.65473 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.04 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7326609 98 - 22422827 GUCCUACCCCUACCCCUUUUCCCCCUGCGCCGCCCUUUGCCCUUGCCAACGAAAUGCGGAUAGCGGGACGCUCUGCGUGUUUGAUUACCACGCGCGGU .......................(((((.((((..((((..........))))..))))...))))).....((((((((.........)))))))). ( -26.30, z-score = -0.98, R) >droSim1.chrX 5853780 90 - 17042790 ---GUCCCCCUACCCC--UUCCCCCUGC---GCCCUUUGCCCUUGCCAACGAAAUGCGGAUAGCGGGACGCUCUGCGUGUUUGAUUACCACGCGCGGU ---(((((.(((.((.--........((---(.....)))....((.........)))).))).)))))...((((((((.........)))))))). ( -24.00, z-score = -0.58, R) >droSec1.super_24 535835 90 - 912625 ---GUCCCCCUACCCC--UUCCCCCUGC---GCCCUUUGCCCUUGCCAACGAAAUGCGGAUAGCGGGACGCUCUGCGUGCUUGAUUACCACGCGCGGU ---(((((.(((.((.--........((---(.....)))....((.........)))).))).)))))...(((((((..((.....))))))))). ( -22.80, z-score = 0.01, R) >droYak2.chrX 7753384 92 + 21770863 ------CACCCCCUUCGACCGCCCCUCCCCCCGCCGCCACUGGGCGCCAUCAUCCAACGAACGCGGUACGCGCUGCGUGUUUGAUUACCACGCGCGGU ------.........(((((((..........(.((((....)))).).((.......))..))))).)).(((((((((.........))))))))) ( -25.80, z-score = 0.26, R) >droEre2.scaffold_4690 16237720 95 - 18748788 ---CACCCCCCGCACCCCCCGCCCGUGCCACCACCCACAUUUGCUUCAACGAAACGCGGAUGGCGGGCCGCUCUGCGUGUUUGAUUACCACGCGCGGU ---......((((....((((((.(((........)))((((((...........)))))))))))).......(((((.........))))))))). ( -30.80, z-score = -0.74, R) >droAna3.scaffold_13117 175094 83 - 5790199 ----------GAUGAGGCUUACCCCCGC----CCUGUCGGCCCAGCCACCCCCUUUCGGCCAGCGGG-CGGACCGCGUGUUUGAUUACCACGCGCGGU ----------..............((((----((.((.((((...............)))).)))))-)))(((((((((.........))))))))) ( -34.46, z-score = -1.38, R) >dp4.chrXL_group1e 11305381 78 - 12523060 ------CCCCACGCAAAUUCUCCCCCACA--------------GCCUGCCAACAGGCGGAUCGCGAGCCACACUGCGUGUUUGAUUACCACGCGCGGU ------..((.(((...............--------------(((((....)))))((....(((((.((.....)))))))....))..))).)). ( -22.40, z-score = -0.94, R) >droPer1.super_14 748445 92 - 2168203 ------CCCCACGCAAAUUCUCCCCCACAGCCUCCCCGAUUCCGCCUGCCAACAGGCGGAUCGCGAGCCACACUGCGUGUUUGAUUACCACGCGCGGU ------.....(((...............(.(((..((((.(((((((....))))))))))).))).).....(((((.........)))))))).. ( -31.50, z-score = -3.41, R) >consensus ______CCCCUACCCC_UUCCCCCCUGC___CCCCUUCGCCCUGGCCAACGAAAUGCGGAUAGCGGGACGCUCUGCGUGUUUGAUUACCACGCGCGGU ......................((......................................((((......))))((((.((.....)).)))))). (-10.01 = -10.04 + 0.03)

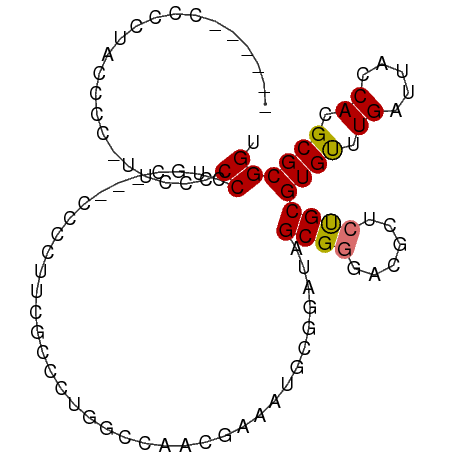

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:24 2011