| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,312,883 – 7,312,988 |
| Length | 105 |
| Max. P | 0.618763 |

| Location | 7,312,883 – 7,312,988 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.92 |
| Shannon entropy | 0.62411 |
| G+C content | 0.55884 |
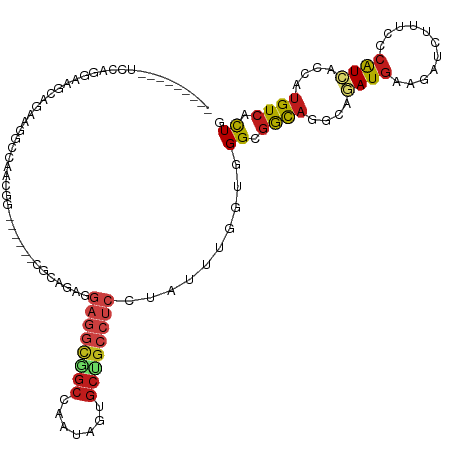
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.41 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
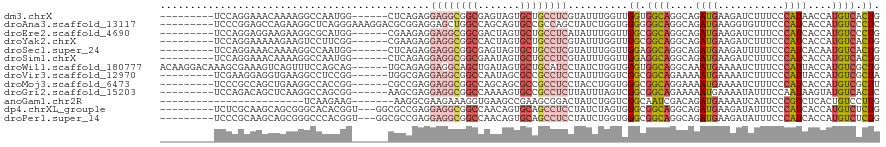
>dm3.chrX 7312883 105 + 22422827 ---------UCCAGGAAACAAAAGGCCAAUGG------CUCAGAGGAGGCGGCGAGUAGUGCUGCCUCGUAUUUGGUUGGUGGCAGGCAGAUGAAGAUCUUUCCCAUAACCAUGUCACUG ---------.((((....)....((((...))------)).....(((((((((.....))))))))).....)))..(((((((((...(((..((....)).)))..)).))))))). ( -33.60, z-score = -0.59, R) >droAna3.scaffold_13117 156947 111 + 5790199 ---------UCCCGGAGCCAGAAGGCUCAGGGAAAGGACGCGGAGGAGCUGGCCAGCAGUGCCGCCAGCUAUCUGGUGGGGGGCAGGCAGAUGAAGGUGUUUCCCAUCACCAUGUCCCUC ---------((((.(((((....))))).))))..(((((.((((((((((((..((...)).))))))).)))((((((..(((..(....)....)))..)))))).)).)))))... ( -54.10, z-score = -2.41, R) >droEre2.scaffold_4690 16224208 105 + 18748788 ---------UCCAGGAGGAAGAAGGCGCAUGG------CGAAGAGGAGGCGGCGACUAGUGCUGCCUCAUAUUUGGUUGGCGGCAGGCAGAUGAAGAUCUUUCCCAUCACCAUGUCCCUG ---------(((....)))...(((.((((((------.(((((((((((((((.....))))))))).((((((.(((....))).)))))).....))))....)).)))))).))). ( -38.90, z-score = -0.97, R) >droYak2.chrX 7738755 105 - 21770863 ---------UCCAGGAAAAAGAAGUCCUUCGG------CGAAGAGGAGGCGGCCACUAGUGCUGCCUCGUAUUUGGUUGGCGGCAGGCAGAUGAAGAUCUUUCCCAUCACCAUGUCACUG ---------...((((........))))((((------(.((((.(((((((((....).)))))))).).))).))))).((((((..((((..((....)).)))).)).)))).... ( -31.80, z-score = 0.22, R) >droSec1.super_24 522185 105 + 912625 ---------UCCAGGAAACAAAAGGCCAAUGG------CUCAGAGGAGGCGGCGAGUAGUGCUGCCUCGUAUUUGGUUGGAGGCAGGCAGAUGAAGAUUUUUCCCAUCACAAUGUCACUG ---------(((((....)....((((...))------))((((.(((((((((.....)))))))))...))))..))))((((....((((..((....)).))))....)))).... ( -32.90, z-score = -0.79, R) >droSim1.chrX 5838758 105 + 17042790 ---------UCCAGGAAACAAAAGGCCAAUGG------CUCAGAGGAGGCGGCGAAUAGUGCUGCCUCGUAUUUGGUUGGAGGCAGGCAGAUGAAGAUCUUUCCCAUCACCAUGUCACUG ---------(((((....)....((((...))------))((((.(((((((((.....)))))))))...))))..))))((((((..((((..((....)).)))).)).)))).... ( -34.30, z-score = -0.86, R) >droWil1.scaffold_180777 1281219 114 - 4753960 ACAAGGACAAAGCGAAAGUCAGUUUCCAGCAG------UGCAGAGGAGGCAGCUGAUAGUGCUGCAUCCUAUCUGGUGGGUGGCAGGCAAAUGAAAAUCUUUCCCAUUACCAUGUCGCUG ....((((...((....))..))))(((.(((------.....((((.(((((.......))))).))))..))).)))((((((((..((((...........)))).)).)))))).. ( -35.10, z-score = -0.49, R) >droVir3.scaffold_12970 3502492 105 + 11907090 ---------UCGAAGGAGGUGAAGGCCUCCGG------UGGCGAGGAGGCGGCCAAUAGCGCCGCCUCCUAUUUGGUCGGCGGCAGAAAAAUGAAAAUCUUUCCCAUUACCAUGUCGCUA ---------.....((((((....))))))((------((((((((((((((((....).))))))))))...((((.(..((..((((.((....)).)))))).).))))))))))). ( -46.00, z-score = -2.92, R) >droMoj3.scaffold_6473 7970197 105 + 16943266 ---------UCCCGCCAGCUGAAGGCCACCGG------CGCCGAGGAGGCGGCCAGCAGCGCCGCCUCCUACCUGGUGGGCGGCAGGAAAAUGAAAAUCUUUCCCAUCACCAUGUCGCUU ---------..(((((((.....((((....)------.))).(((((((((((....).))))))))))..)))))))(((((((((((((....)).)))))........)))))).. ( -48.90, z-score = -2.46, R) >droGri2.scaffold_15203 1616767 105 - 11997470 ---------UCCAGACAGCUCAAGGCCAGCGG------AAGCGAGGAGGCGGCCAAAAGUGCCGCCUCUUAUUUAGUCGGCGGCAGAAAAAUGAAAAUAUUUCCAAUAAGUAUGUCACUC ---------....(((((((....(((.((..------..)).(((((((((((....).))))))))))........)))))).((((.((....)).)))).........)))).... ( -37.80, z-score = -3.63, R) >anoGam1.chr2R 61902628 89 - 62725911 ------------------------UCAAGAAG-------AAGGCGAAGAAAGGUGAAGCCGAAGCGGACUAUCUGGUCGGCAAUCGACAGAUGAAAAUCAUUCCCGUCUCACUGUCCUUG ------------------------........-------((((((..((..((.((((((((..(((.....))).)))))........(((....))).)))))...))..)).)))). ( -23.10, z-score = -0.43, R) >dp4.chrXL_group1e 11290219 108 + 12523060 ---------UCUCGCAAGCAGCGGGCACACGGU---GGCGCCGAGGAGGCGGCCAACAGUGCAGCCUCCUAUCUAGUGGGCGGCAGGCAGAUGAAGAUAUUUCCCAUCACCAUGUCUCUG ---------.(((((.....))))).(((.(((---((.(((((((((((.(((....).)).))))))).((....)).)))).((.(((((....))))).)).))))).)))..... ( -37.90, z-score = 0.18, R) >droPer1.super_14 732653 108 + 2168203 ---------UCCCGCAAGCAGCGGGCCCACGGU---GGCGCCGAGGAGGCGGCCAACAGUGCAGCCUCCUAUCUGGUGGGCGGCAGGCAGAUGAAGAUAUUUCCCAUCACCAUGUCUCUG ---------..((((.....))))((((((((.---....)).(((((((.(((....).)).))))))).....))))))((((((..((((..((....)).)))).)).)))).... ( -45.50, z-score = -0.97, R) >consensus _________UCCAGGAAGCAGAAGGCCAACGG______CGCAGAGGAGGCGGCCAAUAGUGCUGCCUCCUAUUUGGUGGGCGGCAGGCAGAUGAAGAUCUUUCCCAUCACCAUGUCACUG .............................................((((((((.......))))))))..........((.((((....((((...........))))....)))).)). (-15.60 = -16.41 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:21 2011