| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,284,583 – 7,284,689 |
| Length | 106 |
| Max. P | 0.614744 |

| Location | 7,284,583 – 7,284,689 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Shannon entropy | 0.38094 |
| G+C content | 0.41389 |
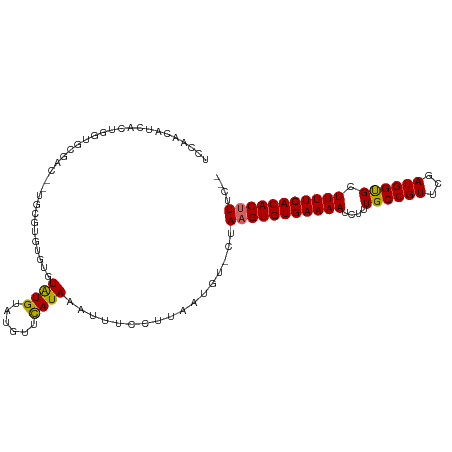
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -15.07 |
| Energy contribution | -14.78 |
| Covariance contribution | -0.29 |
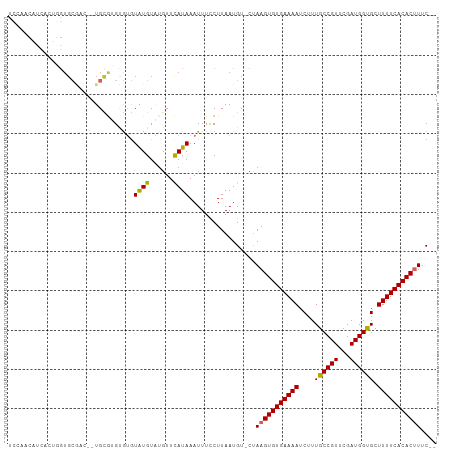
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
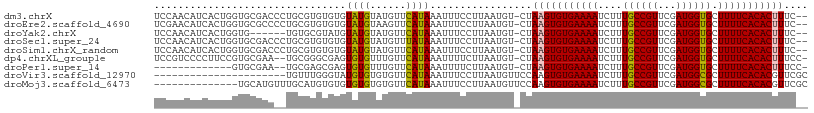
>dm3.chrX 7284583 106 + 22422827 UCCAACAUCACUGGUGCGACCCUGCGUGUGUGUAUGUAUGUUCAUAAAUUUCCUUAAUGU-CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUC-- ....(((((((...((((....))))...))).)))).......................-..(((((((((((.(...(((((...)))))).)))))))))))..-- ( -22.00, z-score = -1.04, R) >droEre2.scaffold_4690 16192617 106 + 18748788 UCGAACAUCACUGGUGCGCCCCUGCGUGUGUGUAUGUAAGUUCAUAAAUUUCCUUAAUGU-CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUC-- ....(((((((..(..(((....)))..)))).)))).......................-..(((((((((((.(...(((((...)))))).)))))))))))..-- ( -24.10, z-score = -1.69, R) >droYak2.chrX 7708330 100 - 21770863 UCCAACAUCACUGGUG------UGUGCGUAUGUAUGUAUGUUCAUAAAUUUCCUUAAUGU-CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUC-- ....((((....((((------((.((((((....)))))).)))).....))...))))-..(((((((((((.(...(((((...)))))).)))))))))))..-- ( -22.00, z-score = -2.01, R) >droSec1.super_24 493857 106 + 912625 UCCAACAUCACUGGUGCGACCCUGCGUGUGUGUAUGUAUGUUUAUAAAUUUCCUUAAUGU-CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUC-- ....(((((((...((((....))))...))).)))).......................-..(((((((((((.(...(((((...)))))).)))))))))))..-- ( -22.00, z-score = -1.23, R) >droSim1.chrX_random 2254948 106 + 5698898 UCCAACAUCACUGGUGCGACCCUGCGUGUGUGUAUGUAUGUUCAUAAAUUUCCUUAAUGU-CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUC-- ....(((((((...((((....))))...))).)))).......................-..(((((((((((.(...(((((...)))))).)))))))))))..-- ( -22.00, z-score = -1.04, R) >dp4.chrXL_group1e 11259592 105 + 12523060 UCCGUCCCCUUCCGUGCGAA--UGCGGGCGAGUGUGUUUGUUCAUAAAUUUUCUUAAUGU-CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUCC- ..((((((.(((.....)))--.).)))))..((((......))))..............-..(((((((((((.(...(((((...)))))).)))))))))))...- ( -23.30, z-score = -1.26, R) >droPer1.super_14 700524 92 + 2168203 -------------GUGCGAA--UGCGAGCGAGUGUGUUUGUUCAUAAAUUUUCUUAAUGU-CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUCC- -------------(.(((((--((((......))))))))).).................-..(((((((((((.(...(((((...)))))).)))))))))))...- ( -20.70, z-score = -1.17, R) >droVir3.scaffold_12970 3466126 87 + 11907090 ----------------------UGUUUGGGUAUGUGUGUGUUCAUAAAUUUCCUUAAUGUUCCAAGUGUGAAAAUCUUUGCCGUUCGAUGGCGCUUUUCACACGUUCGC ----------------------.(((.(((....((((....)))).....))).))).......(((((((((....((((((...)))))).)))))))))...... ( -20.60, z-score = -1.61, R) >droMoj3.scaffold_6473 7930119 95 + 16943266 --------------UGCAUGUUUGCAUGUGUGUGUGUGUGUUCAUAAAUUUCCUUAAUGUUCCAAGUGUGAAAAUCUUUGCCGUUCGAUGGCGCUUUUCACACGUUCGC --------------.(((((..((((..(....)..))))..)))....................(((((((((....((((((...)))))).)))))))))....)) ( -21.60, z-score = -0.74, R) >consensus UCCAACAUCACUGGUGCGAC__UGCGUGUGUGUAUGUAUGUUCAUAAAUUUCCUUAAUGU_CUAAGUGUGAAAAUCUUUGCCGUUCGAUGGUGCUUUUCACACUUUC__ ................................((((......)))).................(((((((((((....((((((...)))))).))))))))))).... (-15.07 = -14.78 + -0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:19 2011