| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,278,255 – 7,278,346 |
| Length | 91 |
| Max. P | 0.744172 |

| Location | 7,278,255 – 7,278,346 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 72.42 |
| Shannon entropy | 0.54205 |
| G+C content | 0.46946 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -13.79 |
| Energy contribution | -14.51 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7278255 91 - 22422827 ----UUGUGUUUGGGUGCUGUGAUCUAUUGUUGGC-AAAGCAAACUUUGCAGCAUACUUGAAGGCGCCACCCACAUACAUACAUACAU---AGCUCUUA ----.(((((.((((((.(((..((...(((((.(-((((....)))))))))).....))..))).)))))).))))).........---........ ( -27.20, z-score = -1.53, R) >droAna3.scaffold_13117 121604 86 - 5790199 --------GUGCGGAGGC--UGAUCUAUUGUUGGC-AGAGC-UACUUUGCUGCAUACCUCGGGGCGCCACCCACAGCGCC-UAUGUGUUUGUGCCUCGA --------.....(((((--............(((-((((.-..)))))))(((.(((...((((((........)))))-)..).)).)))))))).. ( -29.80, z-score = -0.14, R) >droEre2.scaffold_4690 16186577 81 - 18748788 ----UUGUGUUUGGGUGCUGUGAUCUAUUGUUGGUAAAAGCAAACUUUGCAGCAUACUUCAAGGCGCCACCCACAUA--------------AGCUCUUA ----((((((...((((((.(((..(((.((((...((((....)))).)))))))..))).))))))....)))))--------------)....... ( -22.70, z-score = -0.98, R) >droYak2.chrX 7701695 94 + 21770863 ----UUGUGUUUGGGUGCUGUGAUCUAUUGUUGGC-AAAGCAAACUUUGCAGCAUACUUCAAGGCGCCACCCACACAUACACACACACAUAAGCUCUUA ----.(((((...((((((.(((..(((.((((.(-((((....))))))))))))..))).))))))....)))))...................... ( -27.70, z-score = -1.72, R) >droSec1.super_24 487702 83 - 912625 ----UUGUGUUUGGGUGCUGUGAUCUAUUGUUGGC-AAAGCAAACUUUGCAGCAUACUUGAAGGCGCCACCCACAUACAU-----------AGCUCUUA ----.(((((.((((((.(((..((...(((((.(-((((....)))))))))).....))..))).)))))).))))).-----------........ ( -27.20, z-score = -1.82, R) >droSim1.chrX 5821190 83 - 17042790 ----UUGUGUUUGGGUGCUGUGAUCUAUUGUUGGC-AAAGCAAACUUUGCAGCAUACUUGAAGGCGCCACCCACAUACAU-----------AGCUCUUA ----.(((((.((((((.(((..((...(((((.(-((((....)))))))))).....))..))).)))))).))))).-----------........ ( -27.20, z-score = -1.82, R) >droPer1.super_14 692200 87 - 2168203 UUGCGUGGGUGUGGGUGUGCUGAUCCUUUGUUGGC-AAAGCAAACUUUGCCGAAUACUUUGAGGGGCCUCCUCUAGAAUCUUGCGUGU----------- ..(((..((...(((.(.(((...((((..(((((-((((....))))))))).......)))))))).))).......))..)))..----------- ( -26.90, z-score = -0.97, R) >consensus ____UUGUGUUUGGGUGCUGUGAUCUAUUGUUGGC_AAAGCAAACUUUGCAGCAUACUUCAAGGCGCCACCCACAUACAC___________AGCUCUUA .....((((..((((((((((((....(((((......)))))...))))))))).((....))..)))..))))........................ (-13.79 = -14.51 + 0.72)

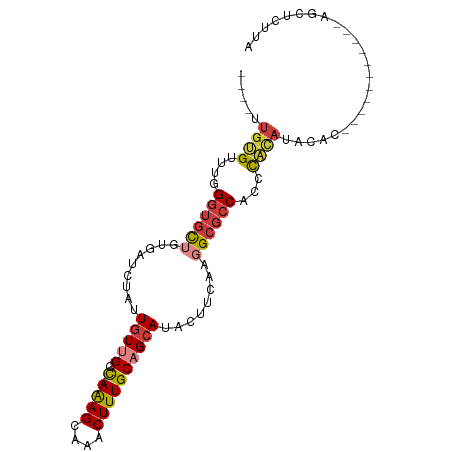
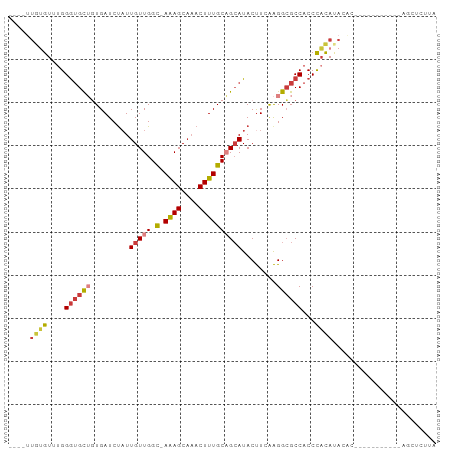
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:16 2011