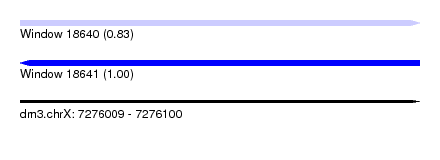
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,276,009 – 7,276,100 |
| Length | 91 |
| Max. P | 0.999517 |

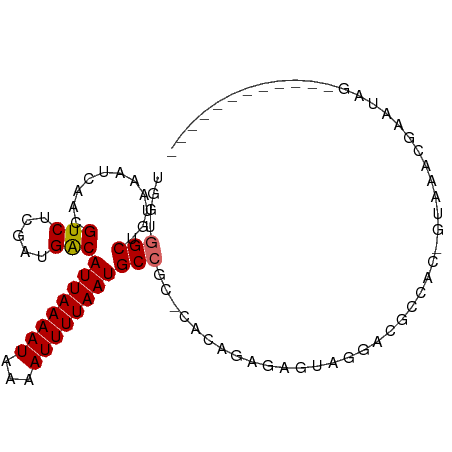
| Location | 7,276,009 – 7,276,100 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 68.56 |
| Shannon entropy | 0.64208 |
| G+C content | 0.41905 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -8.06 |
| Energy contribution | -8.19 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
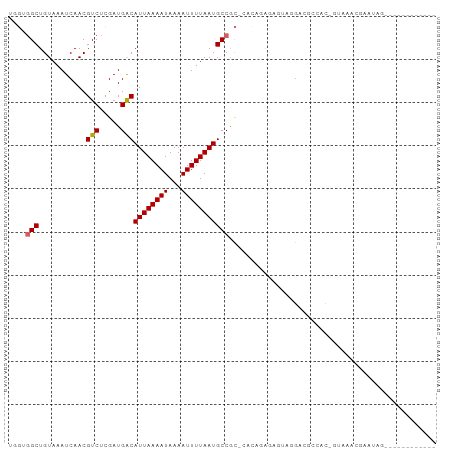
>dm3.chrX 7276009 91 + 22422827 UCGUGGCUGUAAAUCAACGUCUCGAUGGCAUUAAAAUAAAAUUUUAAUGCCGC-CACCGAGAGGAAGACGGCAC-UAAAACGAAUAUUCGUAG------ ..(((.((((...((....(((((.((((((((((((...)))))))))))).-...))))).))..)))))))-....((((....))))..------ ( -26.60, z-score = -3.33, R) >droAna3.scaffold_13117 119094 98 + 5790199 CAGCGGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCGC-CACAAAGGAUAGAAAGCGAUGGGGGGUGGUGGGGCAAGCAGACAG ..((.(((..........(((.....)))((((((((...)))))))).((((-(((........(....)........))))))))))..))...... ( -20.29, z-score = 0.02, R) >droEre2.scaffold_4690 16184186 84 + 18748788 UGGUGGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCGC-CACCGAGAGUAGGACGGCAC--UAAGCGAAUAG------------ .((((((.(((.......(((.....))).(((((((...)))))))))).))-))))..(..(((.......)--))..)......------------ ( -20.60, z-score = -1.75, R) >droYak2.chrX 7699056 84 - 21770863 UGCUGGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCGU-CAGCGAGAGAAGGACGGCAC--UAAACGAAUAG------------ ..(((..(((........(((.....)))..............(((.((((((-(..(....)...))))))).--))))))..)))------------ ( -19.70, z-score = -1.98, R) >droSim1.chrX 5818958 92 + 17042790 UCGUGGCUGUAAAUCAACGUCUUGAUGACAUUAAAAUAAAAUUUUAAUGCCGC-CACCGGGAGAAAGACGACGCACUAAACGAAUACUCGUAG------ ..(((..((.....)).((((((.....(((((((((...)))))))))(((.-...)))....)))))).))).....((((....))))..------ ( -17.60, z-score = -0.69, R) >droVir3.scaffold_12970 8451843 86 - 11907090 UGACUGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCACGCAAAAACGGCACGGCGCUG-CGCCUCCAUUUAG------------ ....((((((........(((.....)))((((((((...))))))))...........)))))).((((...-)))).........------------ ( -16.50, z-score = -0.55, R) >droGri2.scaffold_15203 10424551 87 + 11997470 UGGGUGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCACAAAAAAGAUGUGCGAGGCUGGCGUCUCAGUUCAG------------ ((((((((((......))((((((....(((((((((...))))))))).((((.......)))))))))).)))).))))......------------ ( -20.90, z-score = -1.62, R) >dp4.chrXL_group1e 11249043 72 + 12523060 CAGAGGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCAC-GACAGAG---AGGGAGCAAUGG----------------------- ((...((((.....)..(.((((..((.(((((((((...))))))))).)).-....)))---).).)))..)).----------------------- ( -12.80, z-score = -1.09, R) >droPer1.super_14 689875 72 + 2168203 CAGAGGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCAC-GACAGAG---AGGGAGCAAUGG----------------------- ((...((((.....)..(.((((..((.(((((((((...))))))))).)).-....)))---).).)))..)).----------------------- ( -12.80, z-score = -1.09, R) >consensus UGGUGGCUGUAAAUCAACGUCUCGAUGACAUUAAAAUAAAAUUUUAAUGCCGC_CACAGAGAGUAGGACGCCAC_GUAAACGAAUAG____________ ....(((...........(((.....)))((((((((...)))))))))))................................................ ( -8.06 = -8.19 + 0.12)

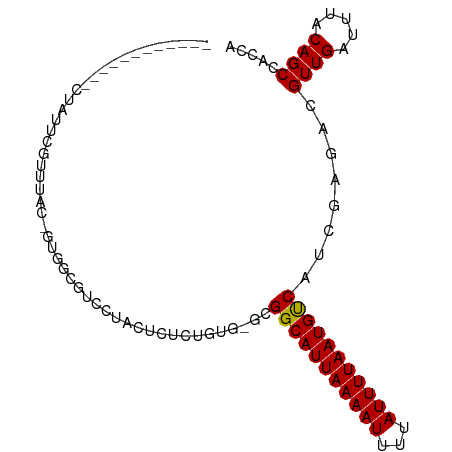
| Location | 7,276,009 – 7,276,100 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 68.56 |
| Shannon entropy | 0.64208 |
| G+C content | 0.41905 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -11.26 |
| Energy contribution | -11.17 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7276009 91 - 22422827 ------CUACGAAUAUUCGUUUUA-GUGCCGUCUUCCUCUCGGUG-GCGGCAUUAAAAUUUUAUUUUAAUGCCAUCGAGACGUUGAUUUACAGCCACGA ------....((((...(((((..-(((((..(........)..)-))(((((((((((...)))))))))))))..)))))...)))).......... ( -25.50, z-score = -2.74, R) >droAna3.scaffold_13117 119094 98 - 5790199 CUGUCUGCUUGCCCCACCACCCCCCAUCGCUUUCUAUCCUUUGUG-GCGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCCGCUG ..........................................(((-(((((((((((((...)))))))))))(((((....))))).....))))).. ( -18.80, z-score = -1.50, R) >droEre2.scaffold_4690 16184186 84 - 18748788 ------------CUAUUCGCUUA--GUGCCGUCCUACUCUCGGUG-GCGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCCACCA ------------......(((((--((((((((((......)).)-)))))))))).....((..((((((((.....))))))))..))..))..... ( -25.00, z-score = -3.61, R) >droYak2.chrX 7699056 84 + 21770863 ------------CUAUUCGUUUA--GUGCCGUCCUUCUCUCGCUG-ACGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCCAGCA ------------.......((((--((((((((...........)-)))))))))))....((..((((((((.....))))))))..))......... ( -23.00, z-score = -3.50, R) >droSim1.chrX 5818958 92 - 17042790 ------CUACGAGUAUUCGUUUAGUGCGUCGUCUUUCUCCCGGUG-GCGGCAUUAAAAUUUUAUUUUAAUGUCAUCAAGACGUUGAUUUACAGCCACGA ------..(((((((((.....))))).))))..........(((-(((((((((((((...)))))))))))(((((....))))).....))))).. ( -25.00, z-score = -2.57, R) >droVir3.scaffold_12970 8451843 86 + 11907090 ------------CUAAAUGGAGGCG-CAGCGCCGUGCCGUUUUUGCGUGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCAGUCA ------------..((((((.((((-...))))...))))))(((.(((((((((((((...))))))))))))))))(((((((.....)))).))). ( -27.80, z-score = -2.65, R) >droGri2.scaffold_15203 10424551 87 - 11997470 ------------CUGAACUGAGACGCCAGCCUCGCACAUCUUUUUUGUGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCACCCA ------------(((....(((.(....).)))...((.(.((((.(((((((((((((...))))))))))))).)))).).)).....)))...... ( -17.60, z-score = -1.42, R) >dp4.chrXL_group1e 11249043 72 - 12523060 -----------------------CCAUUGCUCCCU---CUCUGUC-GUGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCCUCUG -----------------------............---.....((-(((((((((((((...)))))))))))).)))((.((((.....)))).)).. ( -16.20, z-score = -3.23, R) >droPer1.super_14 689875 72 - 2168203 -----------------------CCAUUGCUCCCU---CUCUGUC-GUGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCCUCUG -----------------------............---.....((-(((((((((((((...)))))))))))).)))((.((((.....)))).)).. ( -16.20, z-score = -3.23, R) >consensus ____________CUAUUCGUUUAC_GUGGCGUCCUACUCUCUGUG_GCGGCAUUAAAAUUUUAUUUUAAUGUCAUCGAGACGUUGAUUUACAGCCACCA ................................................(((((((((((...)))))))))))........((((.....))))..... (-11.26 = -11.17 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:15 2011