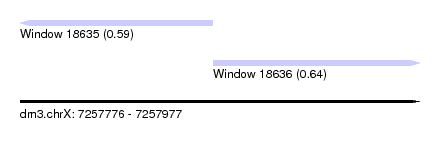
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,257,776 – 7,257,977 |
| Length | 201 |
| Max. P | 0.639528 |

| Location | 7,257,776 – 7,257,873 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.93 |
| Shannon entropy | 0.44432 |
| G+C content | 0.36514 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -10.50 |
| Energy contribution | -10.20 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7257776 97 - 22422827 AAGGCGACCGUCGGUCGUCGAAAAAUGCUAAUUUAUAUA--UAGAUAGAUGUACUCUCUCGCCUUCUGUAUAUAUGUAUAUAU-UGUAGUAAUAAGUAAA ..(((((((...)))))))......(((((...((((((--((.....((((((.............)))))).)))))))).-..)))))......... ( -23.92, z-score = -2.46, R) >droEre2.scaffold_4690 16165513 98 - 18748788 AAGGCGACCGUCGGUCGUCCAAAAAUGCUAAUUUAUAUA--UAGAUAGAUGUACUCUCUCGCCGCUCGUAUAUAUGUACAUAUAUAUAGUAAUAAGUAAA ..(((((((...)))))))......((((...(((((((--((.(((.((((((.............)))))).)))...))))))))).....)))).. ( -21.32, z-score = -1.36, R) >droYak2.chrX 7678828 86 + 21770863 AAGGCGACCGUCGGUCGUCCAAAAAUGCUAAUUUAUAUA--UAGAUAGAUGUACUCACUCGCUCUCCCAUUUUA------------UAGUAAUAAGUAAA ..(((((((...)))))))......((((.(((....((--((((.(((.((........))))).....))))------------))..))).)))).. ( -14.40, z-score = -0.74, R) >droSec1.super_24 467374 86 - 912625 AAGGCGACCGUCGGUCGUCGAAAAAUGCUAAUUUAUAUA--UAGAUAGAUGUACUCUCUCGCCGUCAGUAUAUA------------UAGUAAUAAGUAAA ..(((((((...)))))))......(((((...((((((--(.((..(.((........)))..)).)))))))------------)))))......... ( -19.10, z-score = -1.51, R) >droSim1.chrX 5794764 86 - 17042790 AAGGCGACCGUCGGUCGUCGAAAAAUGCUAAUUUAUAUA--UAGAUAGAUGUACUCUCUCGCCGUCAGUAUAUA------------UAGUAAUAAGUAAA ..(((((((...)))))))......(((((...((((((--(.((..(.((........)))..)).)))))))------------)))))......... ( -19.10, z-score = -1.51, R) >dp4.chrXL_group1e 11227871 75 - 12523060 AAGGCGACCGUCGGUCGCACAAAAAUGCUAAUUUAUAUA--UAGAUAGAUGUACUCGU-----------UUAUA------------UAGUAAUAAGUAAU ...((((((...)))))).......((((.(((..((((--(((((((.....)).))-----------)))))------------))..))).)))).. ( -17.90, z-score = -2.51, R) >droAna3.scaffold_13117 101354 90 - 5790199 GAGGCGACCGACGGUCGUCC-GAAAUGCUAAUUUAUAUAUCUGUAUAGGGAUAGCUAUUCGCCG-CUGUAUGUACUUC------CGUGGAAAUGCGGA-- ..(((((((...)))))))(-(((..((((.(((((((....)))))))..))))..))))(((-(.((.(.(((...------.))).).)))))).-- ( -26.30, z-score = -0.73, R) >consensus AAGGCGACCGUCGGUCGUCCAAAAAUGCUAAUUUAUAUA__UAGAUAGAUGUACUCUCUCGCCGUCAGUAUAUA____________UAGUAAUAAGUAAA ..(((((((...)))))))......((((((((((......))))).((.....))..............................)))))......... (-10.50 = -10.20 + -0.30)


| Location | 7,257,873 – 7,257,977 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.11 |
| Shannon entropy | 0.66392 |
| G+C content | 0.42183 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -11.04 |
| Energy contribution | -10.07 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7257873 104 + 22422827 GGAAUGGUUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUGGCACAUUCCGAGAGAA---GCGGGAGUGAGGUUCGUUUUUGGCCGAAGGUU ...............((((....(((((((...........((((((((......))))))))..(((....---((....))....))))))))))))))...... ( -22.50, z-score = -0.48, R) >droEre2.scaffold_4690 16165611 90 + 18748788 GGAAUGGUUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUGGCACAUUCCGAAAGAA---GUGGGAGUGGGGCUCACU-------------- (((((.((((.(((((.....)))))))))..............(((((......))))))))))......(---(((((.......))))))-------------- ( -20.20, z-score = -1.54, R) >droYak2.chrX 7678914 107 - 21770863 GGAAUGGUUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUGGCACAUUCCAUAAGAAAAUGUGCGAGUGGGUCUCACUUUUUUUUUGGGGUU ......((((.(((((.....)))))))))...........((((((((......))))))))(((.(((((((.(((.((.....)).)))))))))).))).... ( -22.80, z-score = -0.91, R) >dp4.chrXL_group1e 11227946 104 + 12523060 GGAAUGGUUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUGGCAUAUUCCAAGCA-AUGGGGAAGGGGG-UGCUUUCAGUGGGUGACGGGG- (((((.((((.(((((.....)))))))))..............(((((......))))))))))...((-....(((((....-..)))))..))(....)....- ( -18.20, z-score = 0.56, R) >droPer1.super_14 668752 105 + 2168203 GGAAUGGUUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUGGCAUAUUCCAAGCA-AUGGGGAAGGGGGGUGCUUUCAGUGGGUGACGGGG- ...............(.((..(((.((((((..........((((((((......))))))))(((....-.)))..........)))))).)))..)).).....- ( -18.00, z-score = 0.71, R) >droWil1.scaffold_180777 3539706 102 + 4753960 GGAAUGGUUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUAGCACAUUUUACGCAAU--GAUAGGGGCGGGAUGAGGGGCAGAUUGACU--- .....((((.(((....((...(((...............(((((((((......)))))))))..(((..(--....)..)))...)))...)).))).))))--- ( -17.50, z-score = -0.78, R) >droAna3.scaffold_13117 101444 89 + 5790199 GGAAUGGCUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACUUGGCGCAUUCCCGGGGCGUGGGGGCAGUGGGGGCA------------------ ......((((.(((((.....)))))))))...........((((((((......))))))))((((..((......))..))))....------------------ ( -23.10, z-score = -0.35, R) >droVir3.scaffold_12970 3427442 101 + 11907090 GGAAUGCGUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUAGCACAUUUUACGCGG---AACAGCGGC-CCUCAGCCCCUAGGCCAAGUG-- .....(((((.(((((.....))))).)))..........(((((((((......)))))))))...))((---...((.(((-.....))).))...)).....-- ( -23.50, z-score = -0.25, R) >droMoj3.scaffold_6473 7886335 92 + 16943266 GGAAUGCGUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUAGCACAUUUUACGCGG---CACAGGCCA-CGCCCACCUCCG----------- (((..(((((.(((((.....))))).)))..........(((((((((......)))))))))...))((---(....))).-........))).----------- ( -23.80, z-score = -1.77, R) >droGri2.scaffold_15203 1548136 97 - 11997470 GGAAUGCGUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUAGCACAUUUUACGCGG---AACAG-----CGCCCAUCCCCAGUGGAACUU-- ((..(((....(((((.....)))))..))).........(((((((((......)))))))))..(((..---....)-----)).)).(((.....)))....-- ( -20.10, z-score = -0.33, R) >consensus GGAAUGGUUUAUUAAUGGCAAAUUAAAAGCAAUAAAAAACAGAUGUGCUUAACGUGGCACAUUCCAAGCGAA__GGCAGGGGGGGGGCUAUCUCUAGGU_AAGGG__ (((((......(((((.....)))))..................(((((......)))))))))).......................................... (-11.04 = -10.07 + -0.97)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:11 2011