| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,249,691 – 7,249,797 |
| Length | 106 |
| Max. P | 0.887103 |

| Location | 7,249,691 – 7,249,797 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.86 |
| Shannon entropy | 0.70857 |
| G+C content | 0.50333 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -11.49 |
| Energy contribution | -10.85 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
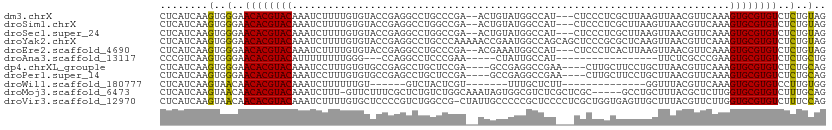
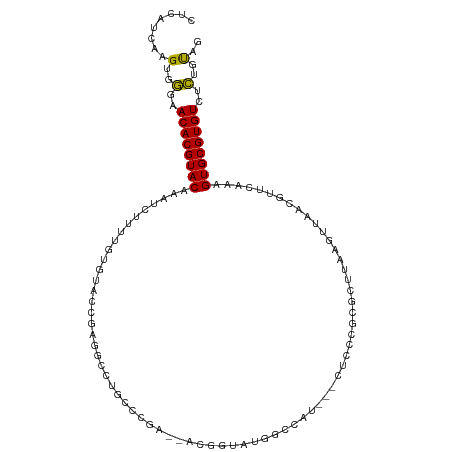
>dm3.chrX 7249691 106 + 22422827 CUCAUCAAGUGGGAACACGUACAAAUCUUUUGUGUACCGAGGCCUGCCCGA--ACUGUAUGGCCAU---CUCCCUCGCUUAAGUUAACGUUCAAAGUGCGUGUCUCUGUAG ........(..(..((((((((....(((..(((....(((((((((....--...))).)))).)---).....)))..)))............))))))))..)..).. ( -25.79, z-score = -0.84, R) >droSim1.chrX 5786396 106 + 17042790 CUCAUCAAGUGGGAACACGUACAAAUCUUUUGUGUACCGAGGCCUGGCCGA--ACUGUAUGGCCAU---CUCCCUCGCUUAAGUUAACGUUCAAAGUGCGUGUCUCUGUAG ........(..(..((((((((...............(((((..((((((.--......)))))).---...))))).....(....).......))))))))..)..).. ( -29.40, z-score = -1.56, R) >droSec1.super_24 459317 106 + 912625 CUCAUCAAGUGGGAACACGUACAAAUCUUUUGUGUACCGAGGCCUGGCCGA--ACUGUAUGGCCAU---CUCCCUCGCUUAAGUUAACGUUCAAAGUGCGUGUCUCUGUAG ........(..(..((((((((...............(((((..((((((.--......)))))).---...))))).....(....).......))))))))..)..).. ( -29.40, z-score = -1.56, R) >droYak2.chrX 7670027 111 - 21770863 CUCAUCAAGUGGGAACACGUACAAAUCUUUUGUGUACCGAGGCCUGCCCAAAAACCGAAUGGCCAGCAGCUCCCGCGCUCAAGUUAACGUUCAAAGUGCGUGUCUCUGUAG ........(..(..((((((((..................((((................))))(((.((....)))))................))))))))..)..).. ( -26.69, z-score = -0.08, R) >droEre2.scaffold_4690 16155863 106 + 18748788 CUCAUCAAGUGGGAACACGUACAAAUCUUUUGUGUACCGAGGCCUGCCCGA--ACGAAAUGGCCAU---CUCCCUCACUUAAGUUAACGUUCAAAGUGCGUGUCUCUGUAG ........(..(..((((((((....(((..(((....((((((.......--.......)))).)---).....)))..)))............))))))))..)..).. ( -24.63, z-score = -0.72, R) >droAna3.scaffold_13117 94042 86 + 5790199 CCCGUCAAGUGGGAACACGUACAUUUUUUUUGGG---CCAGGCCUCCCGAA-----CUAUUGCCAU-----------------UUCUCGCCCGAAGUGCGUGUCUCUGCUG .......((..(..((((((((......((((((---(.(((.....(((.-----...)))....-----------------.))).)))))))))))))))..)..)). ( -28.10, z-score = -2.31, R) >dp4.chrXL_group1e 11213218 103 + 12523060 CUCAUCAAGUGGGAACACGUACAAAUCCUUUGUGUGCCGAGCCUGCUCCGA----GCCGAGGCCGAA----CUUGCUUCCUGCUUAACGUUCAAAGUGCGUGUCUCUGCAG ........(..(..((((((((....((((.((...(.(((....))).).----)).))))..(((----(..((.....)).....))))...))))))))..)..).. ( -31.90, z-score = -1.45, R) >droPer1.super_14 656528 103 + 2168203 CUCAUCAAGUGGGAACACGUACAAAUCCUUUGUGUGCCGAGCCUGCUCCGA----GCCGAGGCCGAA----CUUGCUUCCUGCUUAACGUUCAAAGUGCGUGUCUCUGCAG ........(..(..((((((((....((((.((...(.(((....))).).----)).))))..(((----(..((.....)).....))))...))))))))..)..).. ( -31.90, z-score = -1.45, R) >droWil1.scaffold_180777 3549764 84 + 4753960 CUCAUCAAGUAACAACACGUACAAAUCUUUUUUGU------GUCUACUCGU-------UUUGCUCUU--------------GGUUUACGUUCAAAGUGCGUGUCCUUGUGG ..((.((((.....((((((((...........((------....)).(((-------...((....--------------.))..)))......)))))))).)))))). ( -13.90, z-score = -0.41, R) >droMoj3.scaffold_6473 7873558 105 + 16943266 CUCAUCAAGUAACAACACGUACAAAUCUUU-GUUCUUUCGCUCUGUCUGGCAAAUAGUGGCGUCUCGCUCGC-----GCCUGCUUUACGCUCUUGGUGCGUGUCUUUGCAG ........((((..((((((((........-........(((......)))....((((((((.......))-----))).)))...........))))))))..)))).. ( -25.90, z-score = -1.10, R) >droVir3.scaffold_12970 3416711 110 + 11907090 CUCAUCAAGUAACAACACGUACAAAUCUUUUGUGCUCCCCGUCUGGCCG-CUAUUGCCCCCGCUCCCCUCGCUGGUGAGUUGCUUUACGUUCUUGGUGCGUGUCUUUCCAG (.(((((((.(((.....((((((.....)))))).....((..(((.(-(....))....((((.((.....)).)))).)))..)))))))))))).)........... ( -26.00, z-score = -1.67, R) >consensus CUCAUCAAGUGGGAACACGUACAAAUCUUUUGUGUACCGAGGCCUGCCCGA__ACGGUAUGGCCAU___CUCCCGCGCUUAAGUUAACGUUCAAAGUGCGUGUCUCUGUAG ........(..(..((((((((.........................................................................))))))))..)..).. (-11.49 = -10.85 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:10 2011