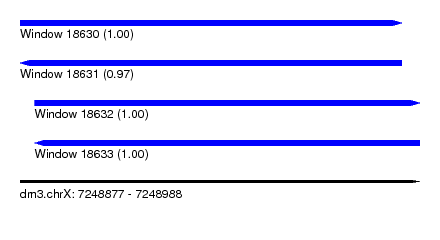
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,248,877 – 7,248,988 |
| Length | 111 |
| Max. P | 0.998944 |

| Location | 7,248,877 – 7,248,983 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.59372 |
| G+C content | 0.48147 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -21.38 |
| Energy contribution | -20.09 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.997246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
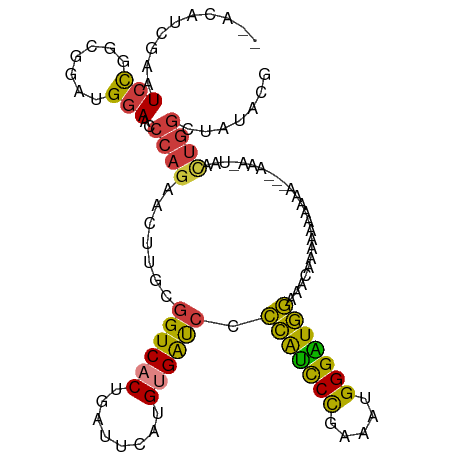
>dm3.chrX 7248877 106 + 22422827 --ACAUCGAAUCCUGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUACCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAA---AAAAUAACUGGCUAUACG --.((((..........))))....((((........(((((........)))))..(((((((.....)))))))..............---.......))))....... ( -25.20, z-score = -2.11, R) >droSim1.chrX 5785560 99 + 17042790 --ACAUCGAAUCCGGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAA----------UAACUGGCUAUACG --........((((.....))))..((((.......((((((........)))))).(((((((.....)))))))...........----------...))))....... ( -27.70, z-score = -1.65, R) >droSec1.super_24 458482 99 + 912625 --ACAUCGAAUCCCGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAA----------UAACUGGCUAUACG --.((((..........))))....((((.......((((((........)))))).(((((((.....)))))))...........----------...))))....... ( -27.20, z-score = -1.89, R) >droYak2.chrX 7667921 91 - 21770863 ----ACCGAGUCUAGCGAAUG-AGCCCAGAACUUGUGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAAUGGAAA--------------- ----.(((..(((.((.....-.))..)))..((((((((((........)))))).(((((((.....)))))))..))))........)))...--------------- ( -25.70, z-score = -2.56, R) >droEre2.scaffold_4690 16155043 107 + 18748788 ----ACCGAAUCCAGCGAACGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAAGAAAAAAUUACUGGCUGUACG ----...........((.((((...((((.......((((((........)))))).(((((((.....)))))))........................)))))))).)) ( -28.00, z-score = -2.52, R) >dp4.chrXL_group1e 11211945 111 + 12523060 AUACAGCAAAUCCGCCGGAUGGAGUCCAGAACUUGCGGUCACUGAUUCAUGCGGCCGGGGCUCUGGUCUGGGGUUCGGAUUUGGGAUACGGGUCCGUUCUUUGGAUAUGGA ......((.((((.(((((((((((((........(((((.(........).)))))))))))).))))))((..((((((((.....))))))))..))..)))).)).. ( -40.80, z-score = -0.83, R) >droPer1.super_14 655147 111 + 2168203 AUACAGCAAAUCCGCCGGAUGGAGUCCAGAACUUGCGGUCACUGAUUCAUGCGGCCGGGGUUCUGGUCUGGGGUUCGGAUUUGGGAUACGGGUCCGUUCUUUGGAUAUGGA ......((.((((.(((((......(((((((((.(((((.(........).)))))))))))))))))))((..((((((((.....))))))))..))..)))).)).. ( -42.70, z-score = -1.58, R) >consensus __ACAUCGAAUCCGGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAA___AAA_UAACUGGCUAUACG ..........(((.......)))..((((.......((((((........)))))).(((((((.....)))))))........................))))....... (-21.38 = -20.09 + -1.30)
| Location | 7,248,877 – 7,248,983 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.59372 |
| G+C content | 0.48147 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7248877 106 - 22422827 CGUAUAGCCAGUUAUUUU---UUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGUAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCAGGAUUCGAUGU-- ......((..((((((..---(((......((..((((((((.....))))))))...))...)))..)))))).)).(((((((((......)).)))))))......-- ( -26.70, z-score = -1.40, R) >droSim1.chrX 5785560 99 - 17042790 CGUAUAGCCAGUUA----------UUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCCGGAUUCGAUGU-- ..............----------.......((..(((((((.....)))))))..)).(((((((((...((.(....).)).((((...))))....))))).))))-- ( -27.30, z-score = -0.53, R) >droSec1.super_24 458482 99 - 912625 CGUAUAGCCAGUUA----------UUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCGGGAUUCGAUGU-- ......((..((((----------((..(((((..(((((((.....)))))))..)).....)))..)))))).))......((((((((......))))))))....-- ( -28.80, z-score = -0.85, R) >droYak2.chrX 7667921 91 + 21770863 ---------------UUUCCAUUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCACAAGUUCUGGGCU-CAUUCGCUAGACUCGGU---- ---------------...((........(((((..(((((((.....)))))))((.((((........))))))))))).(((((.(.-.....))))))...)).---- ( -25.00, z-score = -1.45, R) >droEre2.scaffold_4690 16155043 107 - 18748788 CGUACAGCCAGUAAUUUUUUCUUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCGUUCGCUGGAUUCGGU---- ((.....(((((...................((..(((((((.....)))))))..)).....(((((...((.(....).))..))))).....)))))...))..---- ( -31.60, z-score = -2.02, R) >dp4.chrXL_group1e 11211945 111 - 12523060 UCCAUAUCCAAAGAACGGACCCGUAUCCCAAAUCCGAACCCCAGACCAGAGCCCCGGCCGCAUGAAUCAGUGACCGCAAGUUCUGGACUCCAUCCGGCGGAUUUGCUGUAU ......(((.......)))...((((..((((((((.........(((((((..(((.(((........))).)))...)))))))...((....))))))))))..)))) ( -30.80, z-score = -1.82, R) >droPer1.super_14 655147 111 - 2168203 UCCAUAUCCAAAGAACGGACCCGUAUCCCAAAUCCGAACCCCAGACCAGAACCCCGGCCGCAUGAAUCAGUGACCGCAAGUUCUGGACUCCAUCCGGCGGAUUUGCUGUAU ......(((.......)))...((((..((((((((.........(((((((..(((.(((........))).)))...)))))))...((....))))))))))..)))) ( -30.80, z-score = -2.14, R) >consensus CGUAUAGCCAGUUA_UUU___UUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCCGGAUUCGAUGU__ ...............................(((((((((((.....)))))))))))....((((((...((.(....).)).(((.....)))....))))))...... (-16.14 = -16.70 + 0.56)
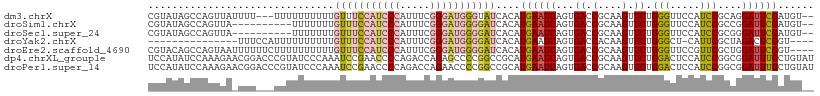
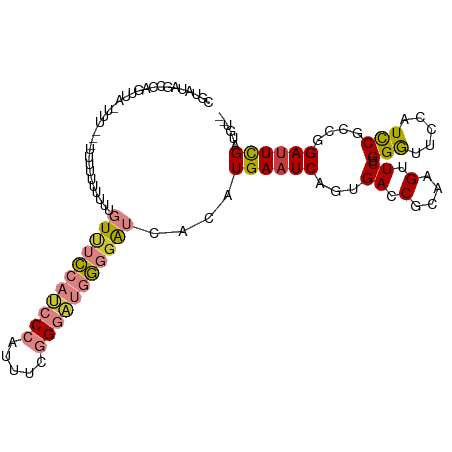
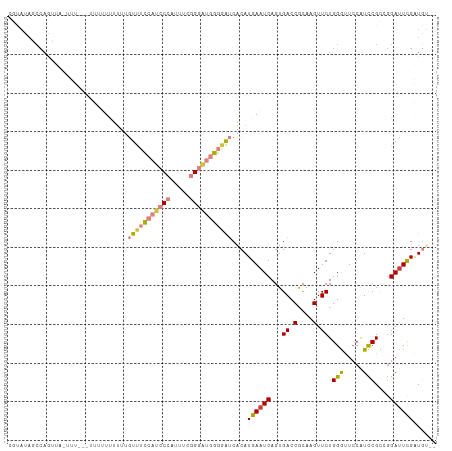
| Location | 7,248,881 – 7,248,988 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.63868 |
| G+C content | 0.47428 |
| Mean single sequence MFE | -32.31 |
| Consensus MFE | -18.73 |
| Energy contribution | -17.03 |
| Covariance contribution | -1.70 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.998944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7248881 107 + 22422827 CGAAUCCUGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUACCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAAAAAAUAACUGGCUAUACGUAUAA----- .......((((.((((...((((........(((((........)))))..(((((((.....))))))).....................)))))))).))))...----- ( -28.00, z-score = -3.07, R) >droSim1.chrX 5785564 100 + 17042790 CGAAUCCGGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAA-------UAACUGGCUAUACGUAUAA----- ........(((.((((...((((.......((((((........)))))).(((((((.....)))))))...........-------...)))))))).)))....----- ( -29.40, z-score = -2.19, R) >droSec1.super_24 458486 100 + 912625 CGAAUCCCGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAA-------UAACUGGCUAUACGUACAA----- ........(((.((((...((((.......((((((........)))))).(((((((.....)))))))...........-------...)))))))).)))....----- ( -29.30, z-score = -2.40, R) >droYak2.chrX 7667923 89 - 21770863 CGAGUCUAGCGAAUG-AGCCCAGAACUUGUGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAAUGG-AAA--------------------- (((((((.((.....-.))..)).))))).((((((........)))))).(((((((.....))))))).................-...--------------------- ( -25.20, z-score = -2.63, R) >droEre2.scaffold_4690 16155045 111 + 18748788 CGAAUCCAGCGAACGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAAGAA-AAAAUUACUGGCUGUACGUAGUAA ........(((.((((...((((.......((((((........)))))).(((((((.....))))))).................-.......)))))))).)))..... ( -30.80, z-score = -2.97, R) >dp4.chrXL_group1e 11211951 111 + 12523060 CAAAUCCGCCGGAUGGAGUCCAGAACUUGCGGUCACUGAUUCAUGCGGCCGGGGCUCUGGUCUGGGGUUCGGAUUUGGGAUACGGGU-CCGUUCUUUGGAUAUGGAUAGAGA ((.((((.(((((((((((((........(((((.(........).)))))))))))).))))))((..((((((((.....)))))-)))..))..)))).))........ ( -40.60, z-score = -0.78, R) >droPer1.super_14 655153 111 + 2168203 CAAAUCCGCCGGAUGGAGUCCAGAACUUGCGGUCACUGAUUCAUGCGGCCGGGGUUCUGGUCUGGGGUUCGGAUUUGGGAUACGGGU-CCGUUCUUUGGAUAUGGAUAGAGA ...(((((...........(((((((((.(((((.(........).))))))))))))))((..(((..((((((((.....)))))-)))..)))..))..)))))..... ( -42.90, z-score = -1.66, R) >consensus CGAAUCCGGCGGAUGGAACCCAGAACUUGCGGUCACUGAUUCAUGUGAUCCCCAUCCCGAAAUGGGAUGGAAACAAAAAAAAAAGG__UAACUGGCUAGACGUACAA_____ ....(((.......))).............((((((........)))))).(((((((.....))))))).......................................... (-18.73 = -17.03 + -1.70)
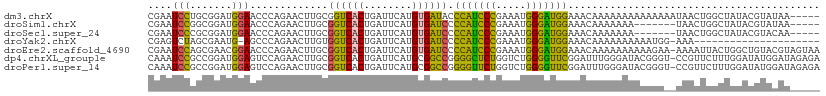
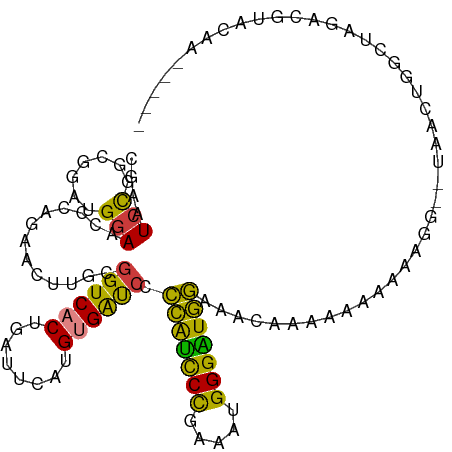
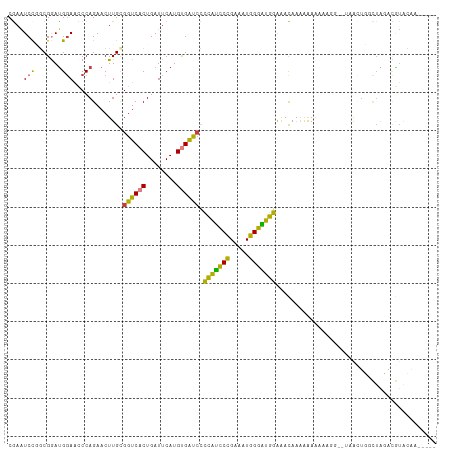
| Location | 7,248,881 – 7,248,988 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.63868 |
| G+C content | 0.47428 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7248881 107 - 22422827 -----UUAUACGUAUAGCCAGUUAUUUUUUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGUAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCAGGAUUCG -----...........((..((((((..(((......((..((((((((.....))))))))...))...)))..)))))).)).(((((((((......)).))))))).. ( -26.70, z-score = -2.22, R) >droSim1.chrX 5785564 100 - 17042790 -----UUAUACGUAUAGCCAGUUA-------UUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCCGGAUUCG -----............((((...-------....(((((..(((((((.....)))))))((.((((........)))))))))))..))))..(((.......))).... ( -26.60, z-score = -0.98, R) >droSec1.super_24 458486 100 - 912625 -----UUGUACGUAUAGCCAGUUA-------UUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCGGGAUUCG -----...........((..((((-------((..(((((..(((((((.....)))))))..)).....)))..)))))).)).......(((((((......))))))). ( -28.70, z-score = -1.16, R) >droYak2.chrX 7667923 89 + 21770863 ---------------------UUU-CCAUUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCACAAGUUCUGGGCU-CAUUCGCUAGACUCG ---------------------...-..........(((((..(((((((.....)))))))((.((((........))))))))))).(((((.(.-.....)))))).... ( -23.60, z-score = -1.72, R) >droEre2.scaffold_4690 16155045 111 - 18748788 UUACUACGUACAGCCAGUAAUUUU-UUCUUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCGUUCGCUGGAUUCG ......((.....(((((......-.............((..(((((((.....)))))))..)).....(((((...((.(....).))..))))).....)))))...)) ( -30.80, z-score = -2.27, R) >dp4.chrXL_group1e 11211951 111 - 12523060 UCUCUAUCCAUAUCCAAAGAACGG-ACCCGUAUCCCAAAUCCGAACCCCAGACCAGAGCCCCGGCCGCAUGAAUCAGUGACCGCAAGUUCUGGACUCCAUCCGGCGGAUUUG ....................(((.-...)))....((((((((.........(((((((..(((.(((........))).)))...)))))))...((....)))))))))) ( -29.60, z-score = -1.88, R) >droPer1.super_14 655153 111 - 2168203 UCUCUAUCCAUAUCCAAAGAACGG-ACCCGUAUCCCAAAUCCGAACCCCAGACCAGAACCCCGGCCGCAUGAAUCAGUGACCGCAAGUUCUGGACUCCAUCCGGCGGAUUUG ....................(((.-...)))....((((((((.........(((((((..(((.(((........))).)))...)))))))...((....)))))))))) ( -29.60, z-score = -2.21, R) >consensus _____AUAUACAUACAGCCAGUUA__CCUUUUUUUUUUGUUUCCAUCCCAUUUCGGGAUGGGGAUCACAUGAAUCAGUGACCGCAAGUUCUGGGUUCCAUCCGCCGGAUUCG ......................................(((((((((((.....))))))))))).....(((((...((.(....).)).(((.....)))....))))). (-16.04 = -16.60 + 0.56)
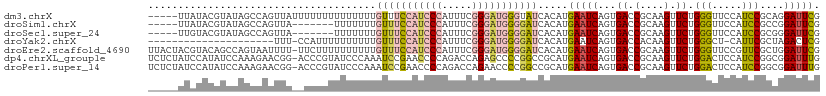
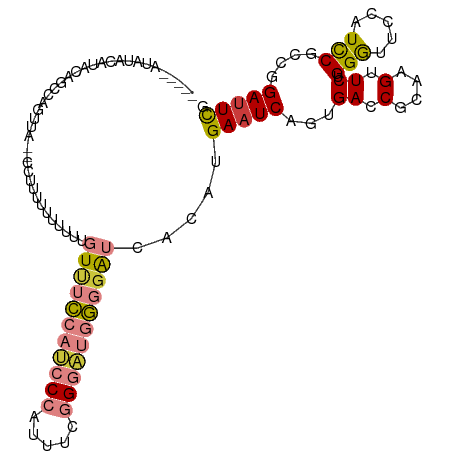
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:09 2011