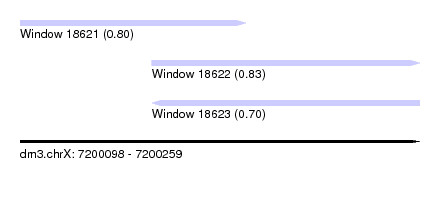
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,200,098 – 7,200,259 |
| Length | 161 |
| Max. P | 0.830948 |

| Location | 7,200,098 – 7,200,189 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 67.22 |
| Shannon entropy | 0.56285 |
| G+C content | 0.40217 |
| Mean single sequence MFE | -10.02 |
| Consensus MFE | -6.42 |
| Energy contribution | -4.18 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.89 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7200098 91 + 22422827 -AUUUUUAUUUUCAUAAAUUUGCAUACACACGCAAGCGAAGCAGCCCACUGUCUCCGACAAAAAAAAAUCACGCACACACAGAACCACACAC -.(((((.((.((.....(((((........))))).((..(((....)))..)).)).)).)))))......................... ( -9.10, z-score = -0.97, R) >droGri2.scaffold_15245 7176013 78 + 18325388 UUUUUGUGUUUUGUUCCCUCCUUUUUUCGACGAGGGCGUGACGACAAGACAUUG--AGUGGAUUUAAAUUAC-CAGAAAAA----------- .(((.(((((((((((((((.((.....)).)))))......)))))))))).)--))(((..........)-))......----------- ( -16.00, z-score = -0.32, R) >droEre2.scaffold_4690 16104640 78 + 18748788 -AUUUUAAUUUUCAUAAAUUUGCAUACACACGCAAGCAAAGCAGCCCACUGUCGCCGACAAAUAAAC-ACACACACACAC------------ -..((((.((.((......((((........))))((...((((....)))).)).)).)).)))).-............------------ ( -6.60, z-score = -0.36, R) >droSec1.super_24 408956 90 + 912625 -AUUUUUAUUUUCAUAAAUUUGCAUACACACGCAAGCGAAGCAGCCCACUGUCUCCGACAAAAAAAA-UCACGCACACACAGAACCACACAC -((((((.((.((.....(((((........))))).((..(((....)))..)).)).)).)))))-)....................... ( -9.20, z-score = -1.12, R) >droSim1.chrX_random 2239813 90 + 5698898 -AUUUUUAUUUUCAUAAAUUUGCAUACACACGCAAGCGAAGCAGCCCACUGUCUCCGACAAAAAAAA-UCACGCACACACAGAACCACACAC -((((((.((.((.....(((((........))))).((..(((....)))..)).)).)).)))))-)....................... ( -9.20, z-score = -1.12, R) >consensus _AUUUUUAUUUUCAUAAAUUUGCAUACACACGCAAGCGAAGCAGCCCACUGUCUCCGACAAAAAAAA_UCACGCACACACAGAACCACACAC ..................(((((........))))).............((((...))))................................ ( -6.42 = -4.18 + -2.24)

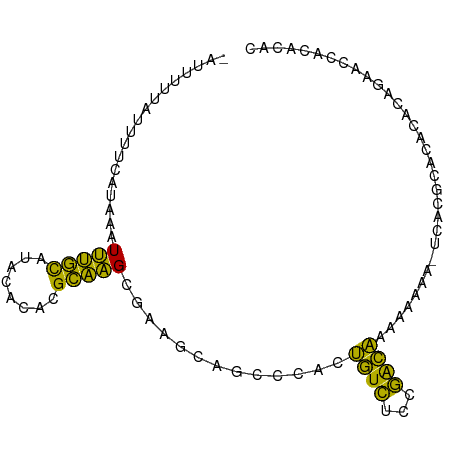
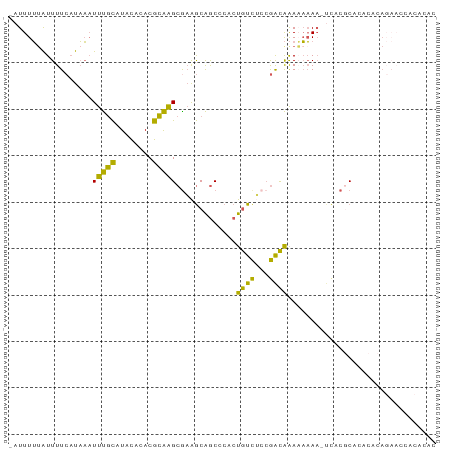
| Location | 7,200,151 – 7,200,259 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Shannon entropy | 0.33255 |
| G+C content | 0.47191 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.50 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.830948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7200151 108 + 22422827 -----CCGACAAAAAAAAAUCACGCACACACAGAACCACACACGCAUGCAAAUAGCGAAAGAGAGCGAGGUGUCUGCGGCAGUGUGUGUGUGUGUUUCAUUCAUGAAU-CCAGA -----...............((((((((((((...((.((.((((.(((......(....)...)))..)))).)).))...)))))))))))).((((....)))).-..... ( -31.00, z-score = -1.24, R) >droEre2.scaffold_4690 16104693 94 + 18748788 ------------------CCGACAAAUAAACACACACACACACGCAUGCGAAUAGCGAAAGAGAGCGAGGUGUUUGCGCCAGUGUGUGCGUGU--UUCAUUCAUGAAUUCCAGA ------------------.........((((((.(((((((.((((.(((....((........))....))).))))...))))))).))))--))................. ( -28.00, z-score = -1.74, R) >droYak2.chrX 7614305 112 - 21770863 CCGACAAAAAAAAACACACACAUACACACCCACAGUCAAACUCGCAUGCGAAUAGCAAAAGAGAGCGAGGUGUGUGCGCCAGUGUGUGUGUGU--UUCAUUCAUGAAUUCCAGA ...........((((((((((((((...............(((...(((.....)))...))).....((((....)))).))))))))))))--))................. ( -33.10, z-score = -2.53, R) >droSec1.super_24 409009 107 + 912625 -----CCGACAAAAAAAA-UCACGCACACACAGAACCACACACGCAUGCAAAUAGCGAAAGAGAGCGAGGUGUCUGCGGCAGUGUGUGUGUGUGUUUCAUUCAUGAAU-CCAGA -----.............-.((((((((((((...((.((.((((.(((......(....)...)))..)))).)).))...)))))))))))).((((....)))).-..... ( -31.00, z-score = -1.18, R) >droSim1.chrX_random 2239866 107 + 5698898 -----CCGACAAAAAAAA-UCACGCACACACAGAACCACACACGCAUGCAAAUAGCGAAAGAGAGCGAGGUGUCUGCGGCAGUGUGUGUGUGUGUUUCAUUCAUGAAU-CCAGA -----.............-.((((((((((((...((.((.((((.(((......(....)...)))..)))).)).))...)))))))))))).((((....)))).-..... ( -31.00, z-score = -1.18, R) >consensus _____CCGACAAAAAAAA_UCACGCACACACAGAACCACACACGCAUGCAAAUAGCGAAAGAGAGCGAGGUGUCUGCGGCAGUGUGUGUGUGUGUUUCAUUCAUGAAU_CCAGA .......................(((((((((....(((((.((((..((....((........))....))..))))...))))))))))))))((((....))))....... (-23.30 = -24.50 + 1.20)


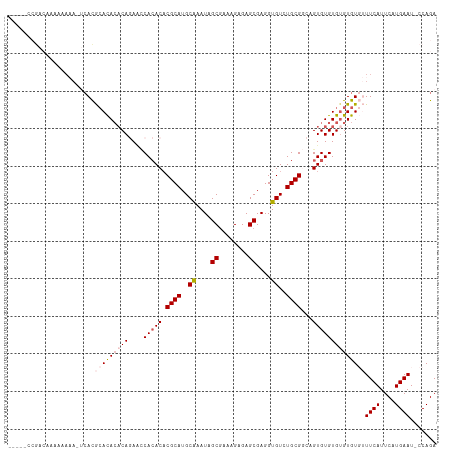
| Location | 7,200,151 – 7,200,259 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Shannon entropy | 0.33255 |
| G+C content | 0.47191 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -19.46 |
| Energy contribution | -21.14 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7200151 108 - 22422827 UCUGG-AUUCAUGAAUGAAACACACACACACACUGCCGCAGACACCUCGCUCUCUUUCGCUAUUUGCAUGCGUGUGUGGUUCUGUGUGUGCGUGAUUUUUUUUUGUCGG----- ....(-((....(((.(((((((.((((((((..((((((.((.....((........)).....(....))).))))))..)))))))).))).)))).))).)))..----- ( -33.30, z-score = -1.90, R) >droEre2.scaffold_4690 16104693 94 - 18748788 UCUGGAAUUCAUGAAUGAA--ACACGCACACACUGGCGCAAACACCUCGCUCUCUUUCGCUAUUCGCAUGCGUGUGUGUGUGUGUUUAUUUGUCGG------------------ ..........(..((((((--.(((((((((((..(((((........((........))........))))))))))))))))))))))..)...------------------ ( -29.69, z-score = -2.31, R) >droYak2.chrX 7614305 112 + 21770863 UCUGGAAUUCAUGAAUGAA--ACACACACACACUGGCGCACACACCUCGCUCUCUUUUGCUAUUCGCAUGCGAGUUUGACUGUGGGUGUGUAUGUGUGUGUUUUUUUUUGUCGG ...........(((..(((--(.(((((((((...((((((.(((.(((..(((...(((.....)))...)))..)))..))).)))))).))))))))).))))....))). ( -35.80, z-score = -2.25, R) >droSec1.super_24 409009 107 - 912625 UCUGG-AUUCAUGAAUGAAACACACACACACACUGCCGCAGACACCUCGCUCUCUUUCGCUAUUUGCAUGCGUGUGUGGUUCUGUGUGUGCGUGA-UUUUUUUUGUCGG----- ....(-((....(((.(((.(((.((((((((..((((((.((.....((........)).....(....))).))))))..)))))))).))).-))).))).)))..----- ( -32.90, z-score = -1.70, R) >droSim1.chrX_random 2239866 107 - 5698898 UCUGG-AUUCAUGAAUGAAACACACACACACACUGCCGCAGACACCUCGCUCUCUUUCGCUAUUUGCAUGCGUGUGUGGUUCUGUGUGUGCGUGA-UUUUUUUUGUCGG----- ....(-((....(((.(((.(((.((((((((..((((((.((.....((........)).....(....))).))))))..)))))))).))).-))).))).)))..----- ( -32.90, z-score = -1.70, R) >consensus UCUGG_AUUCAUGAAUGAAACACACACACACACUGCCGCAGACACCUCGCUCUCUUUCGCUAUUUGCAUGCGUGUGUGGUUCUGUGUGUGCGUGA_UUUUUUUUGUCGG_____ .......((((....)))).(((.((((((((..((((((.((.....((........)).....(....))).))))))..)))))))).))).................... (-19.46 = -21.14 + 1.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:23:01 2011