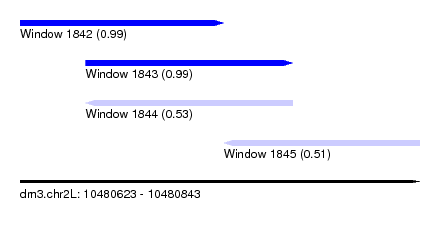
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,480,623 – 10,480,843 |
| Length | 220 |
| Max. P | 0.988582 |

| Location | 10,480,623 – 10,480,735 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Shannon entropy | 0.35050 |
| G+C content | 0.36520 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -20.89 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
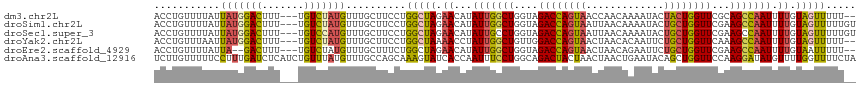
>dm3.chr2L 10480623 112 + 23011544 ACCUGUUUUAUUAUGGACUUU---UGUCUAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACCAACAAAAUACUACUGGUUCGCAGCCAAUUUUGUAGUUUUU-- ...(((((((.(((((((...---.)))))))...(((....)))))))))).(((((((.(..((((((((.............))))))))..))))))))............-- ( -33.32, z-score = -3.36, R) >droSim1.chr2L 10284011 114 + 22036055 ACCUGUUUUAUUAUGGACUUU---UGUCUAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUUGU ...(((((((.(((((((...---.)))))))...(((....)))))))))).(((((((....((((((((.............))))))))...))))))).............. ( -30.52, z-score = -2.47, R) >droSec1.super_3 5908182 114 + 7220098 ACCUGUUUUAUUAUGGACUUU---UGUCCAUGUUUGCUUCCUGGCUAGAACAUAUUGCCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUUGU ...(((((((.(((((((...---.)))))))...(((....)))))))))).(((((.(((....))))))))..((((((.(((((((((.....)))).....))))))))))) ( -30.30, z-score = -2.61, R) >droYak2.chr2L 6886892 112 + 22324452 ACCUGUUUAAUUAUGGACUUU---UGUCUAUGUUUGCUUCCUGGCUAAAACCUAUUGGCUGUUGGACCAGUAACUAACACAAUUCUGCUGGUUCAAAGCCAAUUUUGUAGUUUUU-- .......(((.(((((((...---.))))))).)))......(((((......(((((((.(((((((((((.............))))))))))))))))))....)))))...-- ( -32.42, z-score = -3.71, R) >droEre2.scaffold_4929 11692965 110 - 26641161 ACCUGUUUUAUUA--GACUUU---UGUCUAUGUUUGCUUUCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAGAAUUCUGCUGGUUCGAAGCCAAUUUUGUAAUUUUU-- ...(((((((.((--(((...---.))))).....(((....)))))))))).(((((((....((((((((.............))))))))...)))))))............-- ( -28.72, z-score = -2.47, R) >droAna3.scaffold_12916 14323912 117 + 16180835 UCUUGUUUUUCCUUUGAUCUCAUCUGUUUAUGUUUGCCAGCAAAGUAUCACCAAUUUCCUGGCAGACUACUAACUAACUGAAUACAGCUGGUUCCAAGGAUAUGUUUUGGUUUUCUA .........(((((.((..............(((((((((.(((..........))).))))))))).....((((.(((....))).)))))).)))))................. ( -23.40, z-score = -0.78, R) >consensus ACCUGUUUUAUUAUGGACUUU___UGUCUAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUU__ ...........(((((((.......)))))))..........(((((.((...(((((((....((((((((.............))))))))...))))))).)).)))))..... (-20.89 = -21.04 + 0.15)
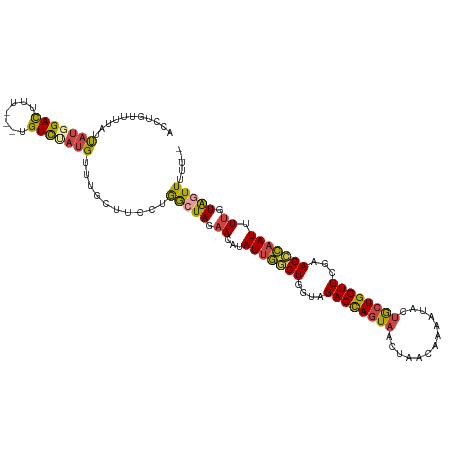
| Location | 10,480,659 – 10,480,773 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.27 |
| Shannon entropy | 0.67304 |
| G+C content | 0.37843 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
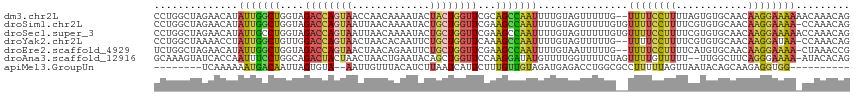
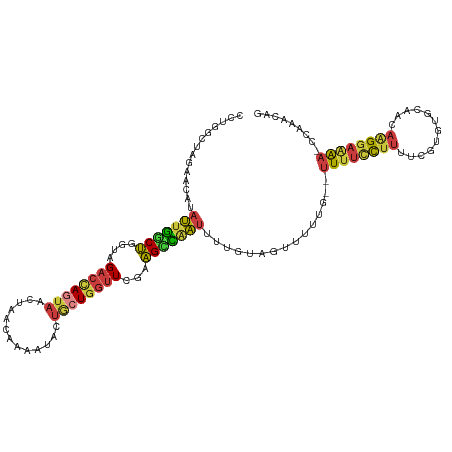
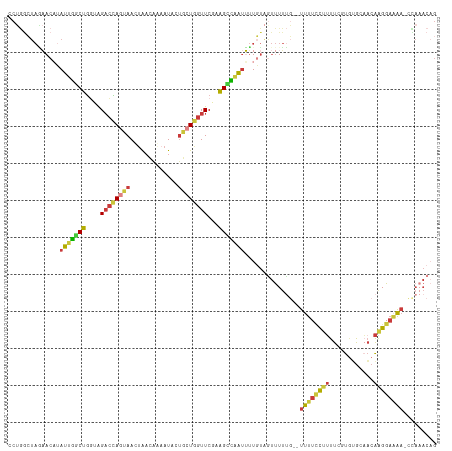
>dm3.chr2L 10480659 114 + 23011544 CCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACCAACAAAAUACUACUGGUUCGCAGCCAAUUUUGUAGUUUUUG--UUUUCCUUUUAGUGUGCAACAAGGAAAAAACAAACAG .(((((((.((...(((((((.(..((((((((.............))))))))..)))))))).)).)))).((((--((((((((...((.....)))))))..)))))))))) ( -33.72, z-score = -2.99, R) >droSim1.chr2L 10284047 115 + 22036055 CCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUUGUGUUUUCCUUUUCGUGUGCAACAAGGAAAA-CCAAACAG .(((((((.((...(((((((....((((((((.............))))))))...))))))).)).)))).((((.(((((((((...((.....))))))))))-)))))))) ( -35.12, z-score = -3.16, R) >droSec1.super_3 5908218 116 + 7220098 CCUGGCUAGAACAUAUUGCCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUUGUGUUUUCCUUUUCGUGUGCAACAAGGAAAAACCAAACAG .(((((((.((...((((.((....((((((((.............))))))))...)).)))).)).)))).((((..((((((((...((.....))))))))))..))))))) ( -25.62, z-score = -0.56, R) >droYak2.chr2L 6886928 113 + 22324452 CCUGGCUAAAACCUAUUGGCUGUUGGACCAGUAACUAACACAAUUCUGCUGGUUCAAAGCCAAUUUUGUAGUUUUUG--UUUUCCUUUUCGUGUGCAACAAGGAUAA-CCAAACAG .(((((((......(((((((.(((((((((((.............))))))))))))))))))....))))(((.(--((.(((((...((.....))))))).))-).)))))) ( -32.72, z-score = -2.79, R) >droEre2.scaffold_4929 11692999 113 - 26641161 UCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAGAAUUCUGCUGGUUCGAAGCCAAUUUUGUAAUUUUUG--UUUUCCUUUUCAUGUGCAACAAGGAAAA-CUAAACCG ...((.....(((.(((((((....((((((((.............))))))))...)))))))..))).......(--((((((((............))))))))-)....)). ( -29.72, z-score = -2.28, R) >droAna3.scaffold_12916 14323951 113 + 16180835 GCAAAGUAUCACCAAUUUCCUGGCAGACUACUAACUAACUGAAUACAGCUGGUUCCAAGGAUAUGUUUUGGUUUUCUAGUUUUGUUUUU--UUGGCUUCAGGGAAAA-AUACACAG .....((((......((((((((.((........((((..(((((.((((((..((((((.....))))))....)))))).)))))..--))))))))))))))..-)))).... ( -22.50, z-score = -0.00, R) >apiMel3.GroupUn 7568001 96 + 399230636 --------UCAAAAAAUGACAAUUAGUGUA--AAUUGUUUACAUCUUAAUCAUUCUUUGUUGUAGAUGAGACCUGGCGCCUUUUUAGUUAAUACAGCAAGAGGUGG---------- --------.........(((((((......--)))))))..(((((....((.....))....)))))........((((((((..(((.....))))))))))).---------- ( -16.80, z-score = 0.03, R) >consensus CCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUUG__UUUUCCUUUUCGUGUGCAACAAGGAAAA_CCAAACAG ..............(((((((....((((((((.............))))))))...)))))))...............((((((((............))))))))......... (-13.57 = -13.72 + 0.15)
| Location | 10,480,659 – 10,480,773 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.27 |
| Shannon entropy | 0.67304 |
| G+C content | 0.37843 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -7.73 |
| Energy contribution | -8.84 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
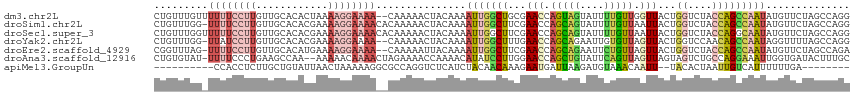
>dm3.chr2L 10480659 114 - 23011544 CUGUUUGUUUUUUCCUUGUUGCACACUAAAAGGAAAA--CAAAAACUACAAAAUUGGCUGCGAACCAGUAGUAUUUUGUUGGUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGG ..((((...((((((((............))))))))--...)))).......(((((((.((((...........((((((...((((....)))))))))).))))))))))). ( -29.50, z-score = -1.61, R) >droSim1.chr2L 10284047 115 - 22036055 CUGUUUGG-UUUUCCUUGUUGCACACGAAAAGGAAAACACAAAAACUACAAAAUUGGCUUCGAACCAGCAGUAUUUUGUUAAUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGG ...(((((-((((((((.(((....))).))))))))).))))..........((((((..((((.(((((....))))).....((((....)))).......)))).)))))). ( -31.80, z-score = -2.86, R) >droSec1.super_3 5908218 116 - 7220098 CUGUUUGGUUUUUCCUUGUUGCACACGAAAAGGAAAACACAAAAACUACAAAAUUGGCUUCGAACCAGCAGUAUUUUGUUAAUUACUGGUCUACCAGGCAAUAUGUUCUAGCCAGG ...((((..((((((((.(((....))).))))))))..))))..........((((((..((((.(((((....))))).....((((....)))).......)))).)))))). ( -30.20, z-score = -2.09, R) >droYak2.chr2L 6886928 113 - 22324452 CUGUUUGG-UUAUCCUUGUUGCACACGAAAAGGAAAA--CAAAAACUACAAAAUUGGCUUUGAACCAGCAGAAUUGUGUUAGUUACUGGUCCAACAGCCAAUAGGUUUUAGCCAGG ....((((-((.(((((.(((....))).)))))...--..((((((.....((((((((((.(((((...(((((...))))).))))).))).))))))).)))))))))))). ( -30.30, z-score = -1.52, R) >droEre2.scaffold_4929 11692999 113 + 26641161 CGGUUUAG-UUUUCCUUGUUGCACAUGAAAAGGAAAA--CAAAAAUUACAAAAUUGGCUUCGAACCAGCAGAAUUCUGUUAGUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGA .......(-((((((((.((......)).))))))))--).............((((((..((((.(((((....))))).....((((....)))).......)))).)))))). ( -30.70, z-score = -2.77, R) >droAna3.scaffold_12916 14323951 113 - 16180835 CUGUGUAU-UUUUCCCUGAAGCCAA--AAAAACAAAACUAGAAAACCAAAACAUAUCCUUGGAACCAGCUGUAUUCAGUUAGUUAGUAGUCUGCCAGGAAAUUGGUGAUACUUUGC ....((((-(.((.((((.((.(..--..................((((.........))))(((.(((((....))))).)))....).))..)))).)).....)))))..... ( -15.80, z-score = 1.41, R) >apiMel3.GroupUn 7568001 96 - 399230636 ----------CCACCUCUUGCUGUAUUAACUAAAAAGGCGCCAGGUCUCAUCUACAACAAAGAAUGAUUAAGAUGUAAACAAUU--UACACUAAUUGUCAUUUUUUGA-------- ----------..((((..((((..............))))..))))...........(((((((((((..((.(((((.....)--))))))....))))))))))).-------- ( -19.14, z-score = -2.19, R) >consensus CUGUUUGG_UUUUCCUUGUUGCACACGAAAAGGAAAA__CAAAAACUACAAAAUUGGCUUCGAACCAGCAGUAUUUUGUUAGUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGG .........((((((((............))))))))...............(((((((...(((.(((((....))))).)))...((....))))))))).............. ( -7.73 = -8.84 + 1.11)
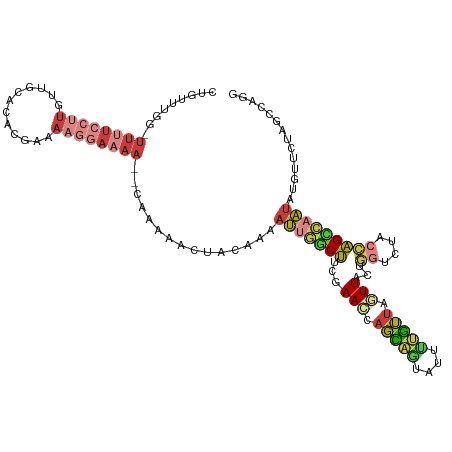
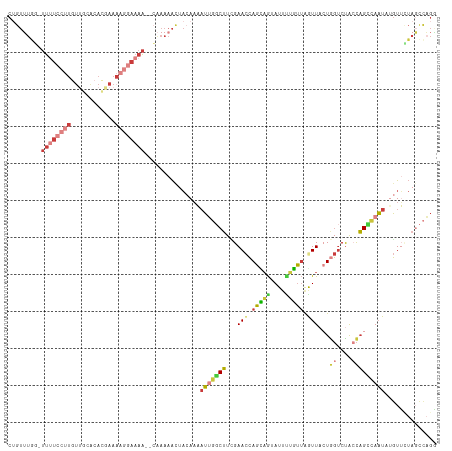
| Location | 10,480,735 – 10,480,843 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.55096 |
| G+C content | 0.40941 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -11.25 |
| Energy contribution | -13.16 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
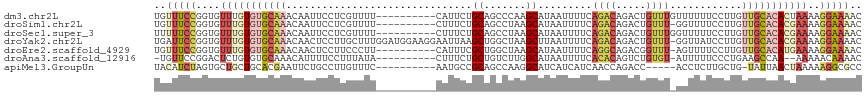
>dm3.chr2L 10480735 108 - 23011544 UGUUUCCGGUGUUUGUGUGCAAACAAUUCCUCGUUUU----------CAUUCUGCAGCCCAAGCAUAAUUUUCAGACAGACUGUUUGUUUUUUCCUUGUUGCACACUAAAAGGAAAAC ..(((((..(....((((((((.(((...........----------.....(((.......))).......(((((.....)))))........)))))))))))..)..))))).. ( -22.70, z-score = -0.62, R) >droSim1.chr2L 10284125 107 - 22036055 UGUUUCCGGUGUUUGUGUGCAAACAAUUCCUCGUUUU----------CUUUCUGCAGCCUAAGCAUAAUUUUCAGACAGACUGUUU-GGUUUUCCUUGUUGCACACGAAAAGGAAAAC ..(((((....(((((((((((.(((...........----------.....(((.......)))......((((((.....))))-))......))))))))))))))..))))).. ( -27.60, z-score = -1.65, R) >droSec1.super_3 5908296 108 - 7220098 UUUUUCCGGUGUUUGUGUGCAAACAAUUCCUCGUUUU----------CUUUCUGCAGCCUAAGCAUAAUUUUCAGACAGACUGUUUGGUUUUUCCUUGUUGCACACGAAAAGGAAAAC .((((((....(((((((((((.(((...........----------.....(((.......)))......((((((.....)))))).......))))))))))))))..)))))). ( -28.20, z-score = -2.08, R) >droYak2.chr2L 6887004 117 - 22324452 UGAUUCCGGUGUUUGUGUGCAAACAACUCCUUGCUUUGGAUGGAAGGAAUUAAGCUGGCUAAGCUUAAUUUUCAGACAGACUGUUU-GGUUAUCCUUGUUGCACACGAAAAGGAAAAC ...((((....(((((((((((.(((.(((.......))).(((..((((((((((.....))))))))))((((((.....))))-))...)))))))))))))))))..))))... ( -40.10, z-score = -3.94, R) >droEre2.scaffold_4929 11693075 107 + 26641161 UGUUUCCGGUGUUUGUGUGCAAACAACUCCUUCCCUU----------CAUUUCGCUGGCUAAGCAUAAUUUUCAGGCAGACGGUUU-AGUUUUCCUUGUUGCACAUGAAAAGGAAAAC ..(((((....((..(((((((.((............----------......(((.....))).........(((.((((.....-.)))).))))))))))))..))..))))).. ( -27.50, z-score = -1.29, R) >droAna3.scaffold_12916 14324029 104 - 16180835 -UGUUCCGGACUCUGUGUGCAAACAUUUUCCUUUAUA----------CUUUCUGCUGUCUUGGCAUAAUUUUCACACAGUCUGUGU-AUUUUUCCCUGAAGCCAA--AAAAACAAAAC -((((..(((....((((....))))..)))......----------............(((((.........((((.....))))-.(((......))))))))--...)))).... ( -14.60, z-score = 0.27, R) >apiMel3.GroupUn 7568065 102 - 399230636 UACAUCUAGUGCUGCUGCACGAAUUCUGCCUUGUUUC----------AAUGCCGCAGCCAAGGCAUCAUCAUCAACCAGACC-----ACCUCUUGCUG-UAUUAACUAAAAAGGCGCC ........((((....)))).......(((((.....----------(((((.(((((....)).............(((..-----...)))))).)-)))).......)))))... ( -21.50, z-score = -0.18, R) >consensus UGUUUCCGGUGUUUGUGUGCAAACAAUUCCUUGUUUU__________CAUUCUGCAGCCUAAGCAUAAUUUUCAGACAGACUGUUU_AGUUUUCCUUGUUGCACACGAAAAGGAAAAC ..(((((....(((((((((((...............................((.......)).........((((.....))))............)))))))))))..))))).. (-11.25 = -13.16 + 1.90)
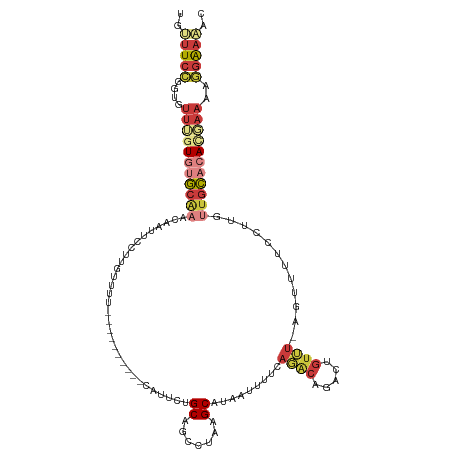
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:20 2011