
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,132,140 – 7,132,252 |
| Length | 112 |
| Max. P | 0.587142 |

| Location | 7,132,140 – 7,132,252 |
|---|---|
| Length | 112 |
| Sequences | 13 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 70.43 |
| Shannon entropy | 0.66371 |
| G+C content | 0.62363 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -10.35 |
| Energy contribution | -11.09 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.88 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
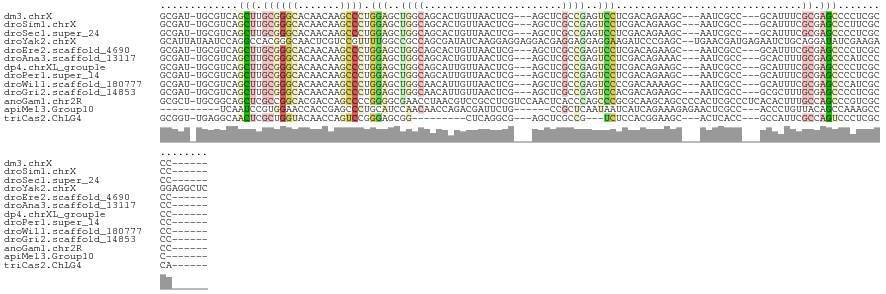
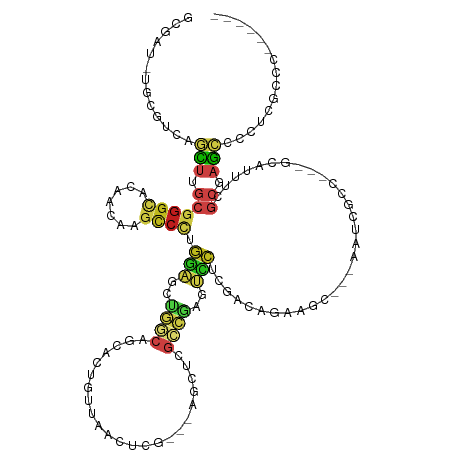
>dm3.chrX 7132140 112 + 22422827 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCACUGUUAACUCG---AGCUCGCCGAGUCCUCGACAGAAGC---AAUCGCC---GCAUUUCGCGAGCCCCUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))..))---)))))).---.......(((((...)))))..------ ( -50.10, z-score = -3.36, R) >droSim1.chrX 5688584 112 + 17042790 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCACUGUUAACUCG---AGCUCGCCGAGUCCUCGACAGAAGC---AAUCGCC---GCAUUUCGCGAGCCCUUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))..))---)))))).---.......(((((...)))))..------ ( -48.10, z-score = -2.76, R) >droSec1.super_24 341101 112 + 912625 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCACUGUUAACUCG---AGCUCGCCGAGUCCUCGACAGAAGC---AAUCGCC---GCAUUUCGCGAGCCCCUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))..))---)))))).---.......(((((...)))))..------ ( -50.10, z-score = -3.36, R) >droYak2.chrX 3488046 126 + 21770863 GCAUUAUAAUCCAGGCCACGGGCAACUCGUCCGUUUUGGCCGCCAGCGAUAUCAAGGAGGAGGACGAGGAGGAGGAAGAUCCCGAGC--UGAACGAUGAGAAUCUGCAGGAUAUCGAAGAGGAGGCUC ((.......(((.((((((((((.....))))))...)))).....(((((((...........((.(((.........)))))..(--((...(((....)))..))))))))))....))).)).. ( -36.60, z-score = 0.34, R) >droEre2.scaffold_4690 16033992 112 + 18748788 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCACUGUUAACUCG---AGCUCGCCGAGUCCUCGACAGAAGC---AAUCGCC---GCAUUUCGCGAGCCCCUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))..))---)))))).---.......(((((...)))))..------ ( -50.10, z-score = -3.36, R) >droAna3.scaffold_13117 2899005 112 + 5790199 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCACUGUUAACUCG---AGCUCGCCGAGUCCUCGACAGAAAC---AAUCGCC---GCACUUUGCGAGCCCAUCCCCC------ (((((-((.((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))...)---))))))(---((.....)))............------ ( -43.40, z-score = -2.63, R) >dp4.chrXL_group1e 11065137 112 + 12523060 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCAUUGUUAACUCG---AGCUCGCCGAGUCCUCGACAGAAGC---AAUCGCC---GCAUUUCGCGAGCCCCUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))..))---)))))).---.......(((((...)))))..------ ( -47.60, z-score = -2.76, R) >droPer1.super_14 502572 112 + 2168203 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCAUUGUUAACUCG---AGCUCGCCGAGUCCUCGACAGAAGC---AAUCGCC---GCAUUUCGCGAGCCCCUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))..))---)))))).---.......(((((...)))))..------ ( -47.60, z-score = -2.76, R) >droWil1.scaffold_180777 1819492 112 + 4753960 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAACAUUGUUAACUCG---AGCUCGCCGAGUCCCCGACAAAAGC---AAUCGCC---GCAUUUCGCGAGCCCAUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....(((((.(((((---.(....))))))....)))))..))---)))))).---.......((((.....))))..------ ( -47.00, z-score = -3.79, R) >droGri2.scaffold_14853 5508469 112 - 10151454 GCGAU-UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAACAUUGUUAACUCG---AGCUCGCCGAGUCCACGACAGAAGC---AAUCGCC---GCGCUUUGCGAGCCCCUCGCCC------ (((((-(((((((((((..((((.......))))..))))))))....((((..(((((---.(....))))))..)))).....))---)))))).---.......(((((...)))))..------ ( -48.10, z-score = -3.03, R) >anoGam1.chr2R 24055121 121 + 62725911 GCGCU-UGCGGCAGCUCGCCGGCACGACCAGCCCCGGGGCGAACCUAACGUCCGCCUCGUCCAACUCACCCAGCCCGCGCAAGCAGCCCCACUCGCCCUCACACUUUGCCAGCCCGUCGCCC------ .....-.(((((.(((.((.(((.......)))..(((((((.....(((.......)))............((..((....)).)).....)))))))........)).)))..)))))..------ ( -34.70, z-score = 0.65, R) >apiMel3.Group10 8129118 102 + 11440700 ----------UCAAUCCGUGGAACCACCGAGCCCUGCAUCCAACAACCAGACGAUUCUG------CCGCUCAAUAAUCAUCAGAAAGAGAACUCGCC---ACCCUGUUACAGCCAAAGCCC------- ----------.......((((.......((((..((.......))..((((....))))------..))))...............((....)).))---))...................------- ( -11.30, z-score = 0.96, R) >triCas2.ChLG4 3789089 100 - 13894384 GCGGU-UGAGGCAACUCGCUGGUACAACCAGUCCGGGAGCGG---------CUCAGGCG---AGCUCGCCG---UCUCCACGGAAGC---ACUCACC---GCCAUUCGCCAGUCCCUCGCCA------ (((((-.(((((......((((.....))))(((((((((((---------(...((..---..)).))))---.)))).)))).))---.))))))---))....................------ ( -39.80, z-score = -1.22, R) >consensus GCGAU_UGCGUCAGCUUGCGGGCACAACAAGCCCUGGAGCUGGCAGCACUGUUAACUCG___AGCUCGCCGAGUCCUCGACAGAAGC___AAUCGCC___GCAUUUCGCGAGCCCCUCGCCC______ .............(((.((((((.......)))).(((..((((.......................))))..)))...............................)).)))............... (-10.35 = -11.09 + 0.74)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:53 2011