| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,092,189 – 7,092,241 |
| Length | 52 |
| Max. P | 0.843211 |

| Location | 7,092,189 – 7,092,241 |
|---|---|
| Length | 52 |
| Sequences | 8 |
| Columns | 53 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
| Shannon entropy | 0.31288 |
| G+C content | 0.43967 |
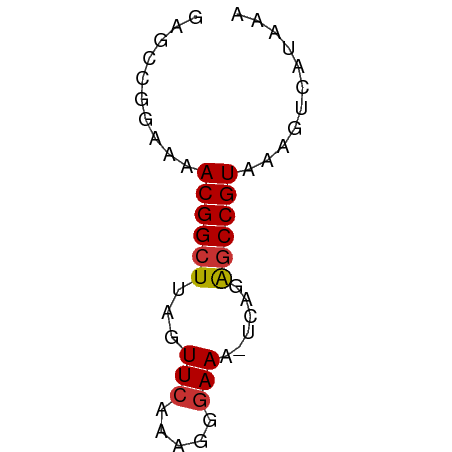
| Mean single sequence MFE | -11.03 |
| Consensus MFE | -7.95 |
| Energy contribution | -7.95 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
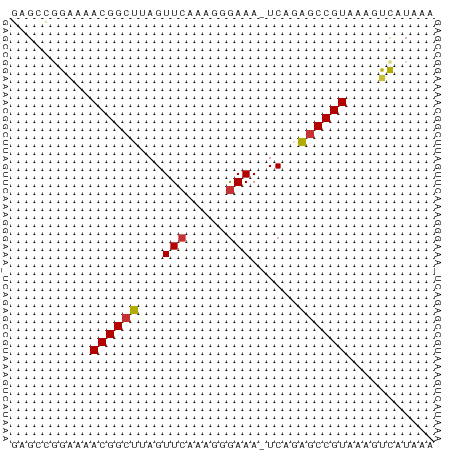
>dm3.chrX 7092189 52 + 22422827 GAGUCGGAAAACGGCUUAGUUCAAAGGGAAA-UCAGAGCCGUAAAGUCAUAAA ......((..(((((((..(((.....))).-...)))))))....))..... ( -11.30, z-score = -2.22, R) >droSim1.chrX 5648868 52 + 17042790 GAGUCGGAAAACGGCUUAGUUCAAAGGGAAA-UCAGGGCCGUAAAGUCAUAAA ......((..(((((((..(((.....))).-...)))))))....))..... ( -10.50, z-score = -1.68, R) >droSec1.super_24 301865 52 + 912625 GAGUCGGAAAACGGCUUAGUUCAAAGGGAAA-UCAGGGCCGUAAAGUCAUAAA ......((..(((((((..(((.....))).-...)))))))....))..... ( -10.50, z-score = -1.68, R) >droYak2.chrX 7498522 52 - 21770863 GAGACGGAAAACGGCUUAGUUCAAAGGGAAA-UCAGGGCCGUAAAGUCAUAAA ..(((.....(((((((..(((.....))).-...)))))))...)))..... ( -13.30, z-score = -2.99, R) >droEre2.scaffold_4690 15993429 52 + 18748788 GAGACGGAAAACGGCUUAGUUCAAAGGGAAA-UCAGGGCCGUAAAGUCAUAAA ..(((.....(((((((..(((.....))).-...)))))))...)))..... ( -13.30, z-score = -2.99, R) >droWil1.scaffold_180777 1773102 52 + 4753960 GCGCUGGGCUACGGCU-AGUUCAAAGUAAAAAUCAGAACCGUAAACCUAUAAA .....((..(((((..-..(((.............))))))))..))...... ( -7.62, z-score = 0.19, R) >droVir3.scaffold_12932 1034365 51 + 2102469 GCGCCUGGUCACGGCU-AGUUCAAAGUGAAA-UCAAAGCCGUAUAACCAUAAA .....((((.((((((-..(((.....))).-....))))))...)))).... ( -12.60, z-score = -2.22, R) >droGri2.scaffold_14853 5454177 51 - 10151454 GCGCCUGAUCACGGCU-AGUUCAAAGCGAAA-UCAAAGCCGUAUAACCAUAAA ..........((((((-..(((.....))).-....))))))........... ( -9.10, z-score = -1.01, R) >consensus GAGCCGGAAAACGGCUUAGUUCAAAGGGAAA_UCAGAGCCGUAAAGUCAUAAA ..........((((((...(((.....)))......))))))........... ( -7.95 = -7.95 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:52 2011